Characterization of recombinant human lactoferrin expressed in Komagataella phaffii
- PMID: 38814097
- PMCID: PMC11215759
- DOI: 10.1039/d4an00333k
Characterization of recombinant human lactoferrin expressed in Komagataella phaffii
Abstract
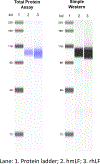
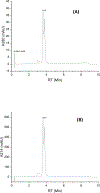
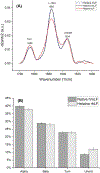
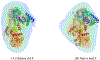
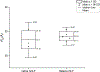
This work presents a thorough characterization of Helaina recombinant human lactoferrin (rhLF, Effera™) expressed in a yeast system at an industrial scale for the first time. Proteomic analysis confirmed that its amino acid sequence is identical to that of native human LF. N-linked glycans were detected at three known glycosylation sites, namely, Asparagines-156, -497, and -642 and they were predominantly oligomannose structures having five to nine mannoses. Helaina rhLF's protein secondary structure was nearly identical to that of human milk lactoferrin (hmLF), as revealed by microfluidic modulation spectroscopy. Results of small-angle X-ray scattering (SAXS) and analytical ultracentrifugation analyses confirmed that, like hmLF, Helaina rhLF displayed well-folded globular structures in solution. Reconstructed solvent envelopes of Helaina rhLF, obtained through the SAXS analysis, demonstrated a remarkable fit with the reported crystalline structure of iron-bound native hmLF. Differential scanning calorimetry investigations into the thermal stability of Helaina rhLF revealed two distinct denaturation temperatures at 68.7 ± 0.9 °C and 91.9 ± 0.5 °C, consistently mirroring denaturation temperatures observed for apo- and holo-hmLF. Overall, Helaina rhLF differed from hmLF in the N-glycans they possessed; nevertheless, the characterization results affirmed that Helaina rhLF was of high purity and exhibited globular structures closely akin to that of hmLF.
Conflict of interest statement
CONFLICTS OF INTEREST
X.L., C.C, A.O., B.H., P.B.B-L, R P., and A J C. are employees of Helaina Inc. N.H.Y. and K.E.W.N. are employees of the Pennsylvania State University. The authors declare no conflict of interests.
Figures


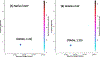
Similar articles
-
Evaluation of the potential food allergy risks of human lactoferrin expressed in Komagataella phaffii.Front Immunol. 2024 Jul 24;15:1380028. doi: 10.3389/fimmu.2024.1380028. eCollection 2024. Front Immunol. 2024. PMID: 39114650 Free PMC article.
-
Workshop report: A study roadmap to evaluate the safety of recombinant human lactoferrin expressed in Komagataella phaffii intended as an ingredient in conventional foods - Recommendations of a scientific expert panel.Food Chem Toxicol. 2024 Aug;190:114817. doi: 10.1016/j.fct.2024.114817. Epub 2024 Jun 14. Food Chem Toxicol. 2024. PMID: 38880466
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
Intravenous magnesium sulphate and sotalol for prevention of atrial fibrillation after coronary artery bypass surgery: a systematic review and economic evaluation.Health Technol Assess. 2008 Jun;12(28):iii-iv, ix-95. doi: 10.3310/hta12280. Health Technol Assess. 2008. PMID: 18547499
Cited by
-
Digestive Profiles of Human Milk, Recombinant Human and Bovine Lactoferrin: Comparing the Retained Intact Protein and Peptide Release.Nutrients. 2024 Jul 21;16(14):2360. doi: 10.3390/nu16142360. Nutrients. 2024. PMID: 39064803 Free PMC article.
-
A Randomized, Double-Blind, Controlled Trial to Assess the Effects of Lactoferrin at Two Doses vs. Active Control on Immunological and Safety Parameters in Healthy Adults.Int J Toxicol. 2025 Jan-Feb;44(1):12-28. doi: 10.1177/10915818241293723. Epub 2024 Oct 28. Int J Toxicol. 2025. PMID: 39465888 Free PMC article. Clinical Trial.
-
Evaluation of the potential food allergy risks of human lactoferrin expressed in Komagataella phaffii.Front Immunol. 2024 Jul 24;15:1380028. doi: 10.3389/fimmu.2024.1380028. eCollection 2024. Front Immunol. 2024. PMID: 39114650 Free PMC article.
-
Four-Week GLP Immunotoxicity Assessment of Lactoferrin Alpha Produced by Komagataella phaffii in Sprague-Dawley Rats.Int J Toxicol. 2025 Mar-Apr;44(2):125-140. doi: 10.1177/10915818241299344. Epub 2024 Nov 13. Int J Toxicol. 2025. PMID: 39537148 Free PMC article.
-
Advancing recombinant protein expression in Komagataella phaffii: opportunities and challenges.FEMS Yeast Res. 2025 Jan 30;25:foaf010. doi: 10.1093/femsyr/foaf010. FEMS Yeast Res. 2025. PMID: 40074550 Free PMC article. Review.
References
-
- Lönnerdal B, Iyer S. Lactoferrin: molecular structure and biological function. Annu Rev Nutr. 1995. Jul;15(1):93–110. - PubMed
-
- Schirmbeck G, Sizonenko S, Sanches E. Neuroprotective role of lactoferrin during early brain development and injury through lifespan. Nutrients [Internet]. 2022. Jul 17 [cited 2023 Apr 4];14(14). Available from: https://pubmed.ncbi.nlm.nih.gov/35889882/ - PMC - PubMed
-
- Wang B, Timilsena YP, Blanch E, Adhikari B. Lactoferrin: structure, function, denaturation and digestion. Critical Reviews in Food Science and Nutrition. 2019. Feb 21;59(4):580–96. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

