Identification of the biological functions and chemo-therapeutic responses of ITGB superfamily in ovarian cancer
- PMID: 38814534
- PMCID: PMC11139846
- DOI: 10.1007/s12672-024-01047-4
Identification of the biological functions and chemo-therapeutic responses of ITGB superfamily in ovarian cancer
Abstract
Background: Patients with ovarian cancer (OC) tend to face a poor prognosis due to a lack of typical symptoms and a high rate of recurrence and chemo-resistance. Therefore, identifying representative and reliable biomarkers for early diagnosis and prediction of chemo-therapeutic responses is vital for improving the prognosis of OC.
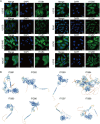
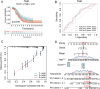
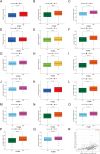
Methods: Expression levels, IHC staining, and subcellular distribution of eight ITGBs were analyzed using The Cancer Genome Atlas (TCGA)-Ovarian Serous Cystadenocarcinoma (OV) database, GEO DataSets, and the HPA website. PrognoScan and Univariate Cox were used for prognostic analysis. TIDE database, TIMER database, and GSCA database were used to analyze the correlation between immune functions and ITGBs. Consensus clustering analysis was performed to subtype OC patients in the TCGA database. LASSO regression was used to construct the predictive model. The Cytoscape software was used for identifying hub genes. The 'pRRophetic' R package was applied to predict chemo-therapeutic responses of ITGBs.
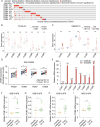
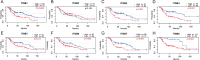
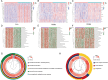
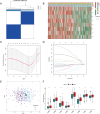
Results: ITGBs were upregulated in OC tissues except ITGB1 and ITGB3. High expression of ITGBs correlated with an unfavorable prognosis of OC except ITGB2. In OC, there was a strong correlation between immune responses and ITGB2, 6, and 7. In addition, the expression matrix of eight ITGBs divided the TCGA-OV database into two subgroups. Subgroup A showed upregulation of eight ITGBs. The predictive model distinguishes OC patients from favorable prognosis to poor prognosis. Chemo-therapeutic responses showed that ITGBs were able to predict responses of common chemo-therapeutic drugs for patients with OC.
Conclusions: This article provides evidence for predicting prognosis, immuno-, and chemo-therapeutic responses of ITGBs in OC and reveals related biological functions of ITGBs in OC.
Keywords: Biological function; Chemo-therapeutic; ITGB; Ovarian cancer; Prognostic prediction; Responses.
© 2024. The Author(s).
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures



Similar articles
-
The prognostic and immune infiltration role of ITGB superfamily members in non-small cell lung cancer.Am J Transl Res. 2022 Sep 15;14(9):6445-6466. eCollection 2022. Am J Transl Res. 2022. PMID: 36247270 Free PMC article.
-
Comprehensive Analysis of the Expression and Prognosis for ITGBs: Identification of ITGB5 as a Biomarker of Poor Prognosis and Correlated with Immune Infiltrates in Gastric Cancer.Front Cell Dev Biol. 2022 Feb 9;9:816230. doi: 10.3389/fcell.2021.816230. eCollection 2021. Front Cell Dev Biol. 2022. PMID: 35223869 Free PMC article.
-
Identification of an Autophagy-Related Signature for Prognosis and Immunotherapy Response Prediction in Ovarian Cancer.Biomolecules. 2023 Feb 9;13(2):339. doi: 10.3390/biom13020339. Biomolecules. 2023. PMID: 36830707 Free PMC article.
-
A novel tumor mutational burden-based risk model predicts prognosis and correlates with immune infiltration in ovarian cancer.Front Immunol. 2022 Aug 8;13:943389. doi: 10.3389/fimmu.2022.943389. eCollection 2022. Front Immunol. 2022. PMID: 36003381 Free PMC article.
-
Identification of STEAP3-based molecular subtype and risk model in ovarian cancer.J Ovarian Res. 2023 Jun 29;16(1):126. doi: 10.1186/s13048-023-01218-x. J Ovarian Res. 2023. PMID: 37386521 Free PMC article.
Cited by
-
Interplay of integrins and selectins in metastasis.Mol Oncol. 2025 Jun;19(6):1582-1611. doi: 10.1002/1878-0261.70026. Epub 2025 May 6. Mol Oncol. 2025. PMID: 40327521 Free PMC article. Review.
References
-
- Eisenhauer EA. Real-world evidence in the treatment of ovarian cancer. Ann Oncol, 2017. 28(suppl_8): p. viii61-viii65. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
