An updated model of shoot apical meristem regulation by ERECTA family and CLAVATA3 signaling pathways in Arabidopsis
- PMID: 38814747
- PMCID: PMC11234387
- DOI: 10.1242/dev.202870
An updated model of shoot apical meristem regulation by ERECTA family and CLAVATA3 signaling pathways in Arabidopsis
Abstract
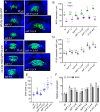
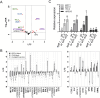
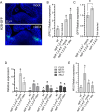
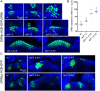
The shoot apical meristem (SAM) gives rise to the aboveground organs of plants. The size of the SAM is relatively constant due to the balance between stem cell replenishment and cell recruitment into new organs. In angiosperms, the transcription factor WUSCHEL (WUS) promotes stem cell proliferation in the central zone of the SAM. WUS forms a negative feedback loop with a signaling pathway activated by CLAVATA3 (CLV3). In the periphery of the SAM, the ERECTA family receptors (ERfs) constrain WUS and CLV3 expression. Here, we show that four ligands of ERfs redundantly inhibit the expression of these two genes. Transcriptome analysis confirmed that WUS and CLV3 are the main targets of ERf signaling and uncovered new ones. Analysis of promoter reporters indicated that the WUS expression domain mostly overlaps with the CLV3 domain and does not shift along the apical-basal axis in clv3 mutants. Our three-dimensional mathematical model captured gene expression distributions at the single-cell level under various perturbed conditions. Based on our findings, CLV3 regulates cellular levels of WUS mostly through autocrine signaling, and ERfs regulate the spatial expression of WUS, preventing its encroachment into the peripheral zone.
Keywords: Arabidopsis; EPFL; ERECTA; Shoot apical meristem; Stem cells; WUSCHEL.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures

Similar articles
-
ERECTA family signaling constrains CLAVATA3 and WUSCHEL to the center of the shoot apical meristem.Development. 2021 Mar 9;148(5):dev189753. doi: 10.1242/dev.189753. Development. 2021. PMID: 33593817
-
Control of Arabidopsis shoot stem cell homeostasis by two antagonistic CLE peptide signalling pathways.Elife. 2021 Oct 13;10:e70934. doi: 10.7554/eLife.70934. Elife. 2021. PMID: 34643181 Free PMC article.
-
HAIRY MERISTEM proteins regulate the WUSCHEL protein levels in mediating CLAVATA3 expression.Physiol Plant. 2024 Sep-Oct;176(5):e14505. doi: 10.1111/ppl.14505. Physiol Plant. 2024. PMID: 39221514
-
WUSCHEL: The essential regulator of the Arabidopsis shoot Apical Meristem.Curr Opin Plant Biol. 2025 Jun;85:102739. doi: 10.1016/j.pbi.2025.102739. Epub 2025 May 16. Curr Opin Plant Biol. 2025. PMID: 40381531 Review.
-
WUSCHEL in the shoot apical meristem: old player, new tricks.J Exp Bot. 2021 Feb 27;72(5):1527-1535. doi: 10.1093/jxb/eraa572. J Exp Bot. 2021. PMID: 33332559 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

