Paramutation at the maize pl1 locus is associated with RdDM activity at distal tandem repeats
- PMID: 38814980
- PMCID: PMC11166354
- DOI: 10.1371/journal.pgen.1011296
Paramutation at the maize pl1 locus is associated with RdDM activity at distal tandem repeats
Abstract
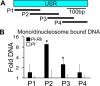
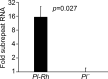
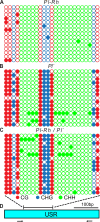
Exceptions to Mendelian inheritance often highlight novel chromosomal behaviors. The maize Pl1-Rhoades allele conferring plant pigmentation can display inheritance patterns deviating from Mendelian expectations in a behavior known as paramutation. However, the chromosome features mediating such exceptions remain unknown. Here we show that small RNA production reflecting RNA polymerase IV function within a distal downstream set of five tandem repeats is coincident with meiotically-heritable repression of the Pl1-Rhoades transcription unit. A related pl1 haplotype with three, but not one with two, repeat units also displays the trans-homolog silencing typifying paramutations. 4C interactions, CHD3a-dependent small RNA profiles, nuclease sensitivity, and polyadenylated RNA levels highlight a repeat subregion having regulatory potential. Our comparative and mutant analyses show that transcriptional repression of Pl1-Rhoades correlates with 24-nucleotide RNA production and cytosine methylation at this subregion indicating the action of a specific DNA-dependent RNA polymerase complex. These findings support a working model in which pl1 paramutation depends on trans-chromosomal RNA-directed DNA methylation operating at a discrete cis-linked and copy-number-dependent transcriptional regulatory element.
Copyright: © 2024 Deans et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: All rmr materials and their uses are covered by U.S. patent 8134047 assigned to The Regents of the University of California. The authors claim no other known competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

