The Toxoplasma gondii F-Box Protein L2 Functions as a Repressor of Stage Specific Gene Expression
- PMID: 38814984
- PMCID: PMC11166348
- DOI: 10.1371/journal.ppat.1012269
The Toxoplasma gondii F-Box Protein L2 Functions as a Repressor of Stage Specific Gene Expression
Abstract
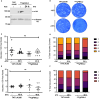
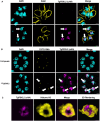
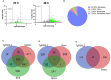
Toxoplasma gondii is a foodborne pathogen that can cause severe and life-threatening infections in fetuses and immunocompromised patients. Felids are its only definitive hosts, and a wide range of animals, including humans, serve as intermediate hosts. When the transmissible bradyzoite stage is orally ingested by felids, they transform into merozoites that expand asexually, ultimately generating millions of gametes for the parasite sexual cycle. However, bradyzoites in intermediate hosts differentiate exclusively to disease-causing tachyzoites, which rapidly disseminate throughout the host. Though tachyzoites are well-studied, the molecular mechanisms governing transitioning between developmental stages are poorly understood. Each parasite stage can be distinguished by a characteristic transcriptional signature, with one signature being repressed during the other stages. Switching between stages require substantial changes in the proteome, which is achieved in part by ubiquitination. F-box proteins mediate protein poly-ubiquitination by recruiting substrates to SKP1, Cullin-1, F-Box protein E3 ubiquitin ligase (SCF-E3) complexes. We have identified an F-box protein named Toxoplasma gondii F-Box Protein L2 (TgFBXL2), which localizes to distinct perinucleolar sites. TgFBXL2 is stably engaged in an SCF-E3 complex that is surprisingly also associated with a COP9 signalosome complex that negatively regulates SCF-E3 function. At the cellular level, TgFBXL2-depleted parasites are severely defective in centrosome replication and daughter cell development. Most remarkable, RNAseq data show that TgFBXL2 conditional depletion induces the expression of stage-specific genes including a large cohort of genes necessary for sexual commitment. Together, these data suggest that TgFBXL2 is a latent guardian of stage specific gene expression in Toxoplasma and poised to remove conflicting proteins in response to an unknown trigger of development.
Copyright: © 2024 Baptista et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Toxoplasma gondii F-Box Protein L2 Silences Feline-Restricted Genes Necessary for Sexual Commitment.bioRxiv [Preprint]. 2023 Dec 18:2023.12.18.572150. doi: 10.1101/2023.12.18.572150. bioRxiv. 2023. PMID: 38187549 Free PMC article. Preprint.
-
Asexual expansion of Toxoplasma gondii merozoites is distinct from tachyzoites and entails expression of non-overlapping gene families to attach, invade, and replicate within feline enterocytes.BMC Genomics. 2015 Feb 13;16(1):66. doi: 10.1186/s12864-015-1225-x. BMC Genomics. 2015. PMID: 25757795 Free PMC article.
-
Toxoplasma F-box protein 1 is required for daughter cell scaffold function during parasite replication.PLoS Pathog. 2019 Jul 26;15(7):e1007946. doi: 10.1371/journal.ppat.1007946. eCollection 2019 Jul. PLoS Pathog. 2019. PMID: 31348812 Free PMC article.
-
Host sensing and signal transduction during Toxoplasma stage conversion.Mol Microbiol. 2021 May;115(5):839-848. doi: 10.1111/mmi.14634. Epub 2020 Nov 21. Mol Microbiol. 2021. PMID: 33118234 Free PMC article. Review.
-
Stress-related and spontaneous stage differentiation of Toxoplasma gondii.Mol Biosyst. 2008 Aug;4(8):824-34. doi: 10.1039/b800520f. Epub 2008 Jun 2. Mol Biosyst. 2008. PMID: 18633484 Review.
Cited by
-
Oxygen-dependent regulation of F-box proteins in Toxoplasma gondii is mediated by Skp1 glycosylation.J Biol Chem. 2024 Nov;300(11):107801. doi: 10.1016/j.jbc.2024.107801. Epub 2024 Sep 21. J Biol Chem. 2024. PMID: 39307307 Free PMC article.
-
Ubiquitin degradation of the Ribonuclease H2 subunit B Rnh2B regulates the virulence of Cryptococcus neoformans.Virulence. 2025 Dec;16(1):2554303. doi: 10.1080/21505594.2025.2554303. Epub 2025 Aug 31. Virulence. 2025. PMID: 40887820 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

