Drug screening in human physiologic medium identifies uric acid as an inhibitor of rigosertib efficacy
- PMID: 38815134
- PMCID: PMC11383364
- DOI: 10.1172/jci.insight.174329
Drug screening in human physiologic medium identifies uric acid as an inhibitor of rigosertib efficacy
Abstract
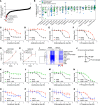
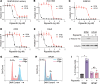
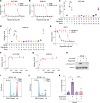
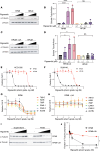
The nonphysiological nutrient levels found in traditional culture media have been shown to affect numerous aspects of cancer cell physiology, including how cells respond to certain therapeutic agents. Here, we comprehensively evaluated how physiological nutrient levels affect therapeutic response by performing drug screening in human plasma-like medium. We observed dramatic nutrient-dependent changes in sensitivity to a variety of FDA-approved and clinically trialed compounds, including rigosertib, an experimental cancer therapeutic that recently failed in phase III clinical trials. Mechanistically, we found that the ability of rigosertib to destabilize microtubules is strongly inhibited by the purine metabolism end product uric acid, which is uniquely abundant in humans relative to traditional in vitro and in vivo cancer models. These results demonstrate the broad and dramatic effects nutrient levels can have on drug response and how incorporation of human-specific physiological nutrient medium might help identify compounds whose efficacy could be influenced in humans.
Keywords: Cancer; Cell biology; Cytoskeleton; Drug screens; Oncology.
Conflict of interest statement
Figures
Update of
-
Drug screening in human physiologic medium identifies uric acid as an inhibitor of rigosertib efficacy.bioRxiv [Preprint]. 2023 Jul 28:2023.07.26.550731. doi: 10.1101/2023.07.26.550731. bioRxiv. 2023. Update in: JCI Insight. 2024 May 30;9(13):e174329. doi: 10.1172/jci.insight.174329. PMID: 37546939 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

