Genomic analysis of Salmonella bacteriophages revealed multiple endolysin ORFs and importance of ligand-binding site of receptor-binding protein
- PMID: 38816206
- PMCID: PMC11180984
- DOI: 10.1093/femsec/fiae079
Genomic analysis of Salmonella bacteriophages revealed multiple endolysin ORFs and importance of ligand-binding site of receptor-binding protein
Abstract
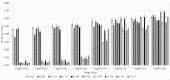
Salmonella is a prevalent foodborne pathogen causing millions of global cases annually. Antimicrobial resistance is a growing public health concern, leading to search for alternatives like bacteriophages. A total of 97 bacteriophages, isolated from cattle farms (n = 48), poultry farms (n = 37), and wastewater (n = 5) samples in Türkiye, were subjected to host-range analysis using 36 Salmonella isolates with 18 different serotypes. The broadest host range belonged to an Infantis phage (MET P1-091), lysing 28 hosts. A total of 10 phages with the widest host range underwent further analysis, revealing seven unique genomes (32-243 kb), including a jumbophage (>200 kb). Except for one with lysogenic properties, none of them harbored virulence or antibiotic resistance genes, making them potential Salmonella reducers in different environments. Examining open reading frames (ORFs) of endolysin enzymes revealed surprising findings: five of seven unique genomes contained multiple endolysin ORFs. Despite sharing same endolysin sequences, phages exhibited significant differences in host range. Detailed analysis unveiled diverse receptor-binding protein sequences, with similar structures but distinct ligand-binding sites. These findings emphasize the importance of ligand-binding sites of receptor-binding proteins. Additionally, bacterial reduction curve and virulence index revealed that Enteritidis phages inhibit bacterial growth even at low concentrations, unlike Infantis and Kentucky phages.
Keywords: Salmonella; antibiotic resistance; bacteriophages; biopreservation; endolysin; ligand-binding site.
© The Author(s) 2024. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
None declared.
Figures
Similar articles
-
Salmonella bacteriophage diversity reflects host diversity on dairy farms.Food Microbiol. 2013 Dec;36(2):275-85. doi: 10.1016/j.fm.2013.06.014. Epub 2013 Jul 4. Food Microbiol. 2013. PMID: 24010608 Free PMC article.
-
Characterization of a Salmonella Enteritidis bacteriophage showing broad lytic activity against Gram-negative enteric bacteria.J Microbiol. 2018 Dec;56(12):917-925. doi: 10.1007/s12275-018-8310-1. Epub 2018 Oct 25. J Microbiol. 2018. PMID: 30361974
-
Phenotypic characterization and genomic analysis of a Salmonella phage L223 for biocontrol of Salmonella spp. in poultry.Sci Rep. 2024 Jul 3;14(1):15347. doi: 10.1038/s41598-024-64999-1. Sci Rep. 2024. PMID: 38961138 Free PMC article.
-
Characterization and therapeutic potential of newly isolated bacteriophages targeting the most common Salmonella serovars in Europe.Sci Rep. 2025 Mar 29;15(1):10872. doi: 10.1038/s41598-025-95398-9. Sci Rep. 2025. PMID: 40157986 Free PMC article.
-
Salmonella phages and prophages--genomics and practical aspects.Methods Mol Biol. 2007;394:133-75. doi: 10.1007/978-1-59745-512-1_9. Methods Mol Biol. 2007. PMID: 18363236 Review.
Cited by
-
Effect of egg cultivation methods on Salmonella prevalence and the promising prevention strategy: bacteriophage therapy.Food Sci Biotechnol. 2025 Mar 27;34(10):2361-2372. doi: 10.1007/s10068-025-01855-6. eCollection 2025 Jun. Food Sci Biotechnol. 2025. PMID: 40351715
-
Layer-by-Layer Coated Alginate Hydrogels with Antibacterial Activity Based on Bacteriophage and Curcumin Combination.ACS Omega. 2025 Jul 23;10(30):33108-33123. doi: 10.1021/acsomega.5c02562. eCollection 2025 Aug 5. ACS Omega. 2025. PMID: 40787330 Free PMC article.
References
-
- Ackermann H-W. Salmonella Phages Examined in the Electron Microscope. Totowa: Humana Press, 2009. - PubMed
-
- Akhtar M, Viazis S, Diez-Gonzalez F. Isolation, identification and characterization of lytic, wide host range bacteriophages from waste effluents against Salmonella enterica serovars. Food Control. 2014;38:67–74. 10.1016/j.foodcont.2013.09.064. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

