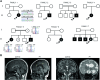
Biallelic variants in CSMD1 are implicated in a neurodevelopmental disorder with intellectual disability and variable cortical malformations
- PMID: 38816421
- PMCID: PMC11140003
- DOI: 10.1038/s41419-024-06768-6
Biallelic variants in CSMD1 are implicated in a neurodevelopmental disorder with intellectual disability and variable cortical malformations
Abstract
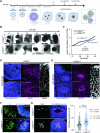
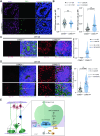
CSMD1 (Cub and Sushi Multiple Domains 1) is a well-recognized regulator of the complement cascade, an important component of the innate immune response. CSMD1 is highly expressed in the central nervous system (CNS) where emergent functions of the complement pathway modulate neural development and synaptic activity. While a genetic risk factor for neuropsychiatric disorders, the role of CSMD1 in neurodevelopmental disorders is unclear. Through international variant sharing, we identified inherited biallelic CSMD1 variants in eight individuals from six families of diverse ancestry who present with global developmental delay, intellectual disability, microcephaly, and polymicrogyria. We modeled CSMD1 loss-of-function (LOF) pathogenesis in early-stage forebrain organoids differentiated from CSMD1 knockout human embryonic stem cells (hESCs). We show that CSMD1 is necessary for neuroepithelial cytoarchitecture and synchronous differentiation. In summary, we identified a critical role for CSMD1 in brain development and biallelic CSMD1 variants as the molecular basis of a previously undefined neurodevelopmental disorder.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Costanzo F, Zanni G, Fucà E, Di Paola M, Barresi S, Travaglini L, et al. Cerebellar Agenesis and Bilateral Polimicrogyria Associated with Rare Variants of CUB and Sushi Multiple Domains 1 Gene (CSMD1): A Longitudinal Neuropsychological and Neuroradiological Case Study. Int J Environ Res Public Health. 2022;19. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

