TCAF1 promotes TRPV2-mediated Ca2+ release in response to cytosolic DNA to protect stressed replication forks
- PMID: 38816425
- PMCID: PMC11139906
- DOI: 10.1038/s41467-024-48988-6
TCAF1 promotes TRPV2-mediated Ca2+ release in response to cytosolic DNA to protect stressed replication forks
Abstract
The protection of the replication fork structure under stress conditions is essential for genome maintenance and cancer prevention. A key signaling pathway for fork protection involves TRPV2-mediated Ca2+ release from the ER, which is triggered after the generation of cytosolic DNA and the activation of cGAS/STING. This results in CaMKK2/AMPK activation and subsequent Exo1 phosphorylation, which prevent aberrant fork processing, thereby ensuring genome stability. However, it remains poorly understood how the TRPV2 channel is activated by the presence of cytosolic DNA. Here, through a genome-wide CRISPR-based screen, we identify TRPM8 channel-associated factor 1 (TCAF1) as a key factor promoting TRPV2-mediated Ca2+ release under replication stress or other conditions that activate cGAS/STING. Mechanistically, TCAF1 assists Ca2+ release by facilitating the dissociation of STING from TRPV2, thereby relieving TRPV2 repression. Consistent with this function, TCAF1 is required for fork protection, chromosomal stability, and cell survival after replication stress.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
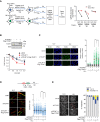
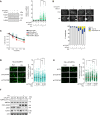
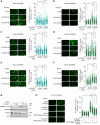
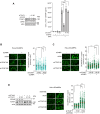


References
MeSH terms
Substances
Grants and funding
- R01 DK115972/DK/NIDDK NIH HHS/United States
- R01 GM098535/GM/NIGMS NIH HHS/United States
- R01 CA193318/CA/NCI NIH HHS/United States
- 5124/WUSTL | Washington University School of Medicine in St. Louis
- 82272984/National Natural Science Foundation of China (National Science Foundation of China)
- R01 CA227001/CA/NCI NIH HHS/United States
- R01CA193318/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- RSG-13-212-01-DMC/American Cancer Society (American Cancer Society, Inc.)
- R01GM098535/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01 DK123301/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

