Real-time reverse transcription recombinase polymerase amplification (RT-RPA) assay for detection of cassava brown streak viruses
- PMID: 38816439
- PMCID: PMC11139904
- DOI: 10.1038/s41598-024-62249-y
Real-time reverse transcription recombinase polymerase amplification (RT-RPA) assay for detection of cassava brown streak viruses
Abstract
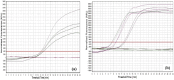
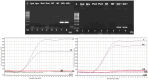
Cassava brown streak disease (CBSD) caused by Cassava brown streak virus (CBSV) and Ugandan cassava brown streak virus (UCBSV) is the most economically important viral disease of cassava. As cassava is a vegetatively propagated crop, the development of rapid and sensitive diagnostics would aid in the identification of virus-free planting material and development of effective management strategies. In this study, a rapid, specific and sensitive real-time reverse transcription recombinase polymerase amplification (RT-RPA) assay was developed for real-time detection of CBSV and UCBSV. The RT-RPA was able to detect as little as 2 pg/µl of purified RNA obtained from infected cassava leaves, a sensitivity equivalent to that obtained by quantitative real-time reverse transcription PCR (qRT-PCR), within 20 min at 37 °C. Further, the RT-RPA detected each target virus directly from crude leaf and stem extracts, avoiding the tedious and costly isolation of high-quality RNA. The developed RT-RPA assay provides a valuable diagnostic tool that can be adopted by cassava seed certification and virus resistance breeding programs to ensure distribution of virus-free cassava planting materials to farmers. This is the first report on the development and validation of crude sap-based RT-RPA assay for the detection of cassava brown streak viruses (UCBSV and CBSV) infection in cassava plants.
Keywords: Cassava brown streak viruses; Early virus detection; Isothermal amplification; Rapid diagnosis; Reverse transcriptase recombinase polymerase amplification (RT-RPA).
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Efficient transmission of cassava brown streak disease viral pathogens by chip bud grafting.BMC Res Notes. 2013 Dec 6;6:516. doi: 10.1186/1756-0500-6-516. BMC Res Notes. 2013. PMID: 24314370 Free PMC article.
-
Simultaneous virus-specific detection of the two cassava brown streak-associated viruses by RT-PCR reveals wide distribution in East Africa, mixed infections, and infections in Manihot glaziovii.J Virol Methods. 2011 Feb;171(2):394-400. doi: 10.1016/j.jviromet.2010.09.024. Epub 2010 Oct 13. J Virol Methods. 2011. PMID: 20923689
-
Loop-mediated isothermal amplification for rapid detection of the causal agents of cassava brown streak disease.J Virol Methods. 2013 Aug;191(2):148-54. doi: 10.1016/j.jviromet.2012.07.015. Epub 2012 Jul 20. J Virol Methods. 2013. PMID: 22820076
-
Cassava brown streak disease: a threat to food security in Africa.J Gen Virol. 2015 May;96(Pt 5):956-68. doi: 10.1099/vir.0.000014. J Gen Virol. 2015. PMID: 26015320 Review.
-
Incidence, severity and distribution of Cassava brown streak disease in northeastern Democratic Republic of Congo.Cogent Food Agric. 2020 Jul 16;6(1):1789422. doi: 10.1080/23311932.2020.1789422. Cogent Food Agric. 2020. PMID: 33718519 Free PMC article. Review.
Cited by
-
Portable solutions for plant pathogen diagnostics: development, usage, and future potential.Front Microbiol. 2025 Jan 31;16:1516723. doi: 10.3389/fmicb.2025.1516723. eCollection 2025. Front Microbiol. 2025. PMID: 39959158 Free PMC article. Review.
References
-
- Robson F, Hird DL, Boa E. Cassava brown streak: A deadly virus on the move. Plant Pathol. 2024;73:221–241. doi: 10.1111/ppa.13807. - DOI
-
- Hillocks R, Jennings D. Cassava brown streak disease: A review of present knowledge and research needs. Int. J. Pest Manag. 2003;49(3):225–234. doi: 10.1080/0967087031000101061. - DOI
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

