AmiA and AliA peptide ligands, found in Klebsiella pneumoniae, are imported into pneumococci and alter the transcriptome
- PMID: 38816440
- PMCID: PMC11139975
- DOI: 10.1038/s41598-024-63217-2
AmiA and AliA peptide ligands, found in Klebsiella pneumoniae, are imported into pneumococci and alter the transcriptome
Abstract
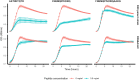
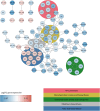
Klebsiella pneumoniae releases the peptides AKTIKITQTR and FNEMQPIVDRQ, which bind the pneumococcal proteins AmiA and AliA respectively, two substrate-binding proteins of the ABC transporter Ami-AliA/AliB oligopeptide permease. Exposure to these peptides alters pneumococcal phenotypes such as growth. Using a mutant in which a permease domain of the transporter was disrupted, by growth analysis and epifluorescence microscopy, we confirmed peptide uptake via the Ami permease and intracellular location in the pneumococcus. By RNA-sequencing we found that the peptides modulated expression of genes involved in metabolism, as pathways affected were mostly associated with energy or synthesis and transport of amino acids. Both peptides downregulated expression of genes involved in branched-chain amino acid metabolism and the Ami permease; and upregulated fatty acid biosynthesis genes but differed in their regulation of genes involved in purine and pyrimidine biosynthesis. The transcriptomic changes are consistent with growth suppression by peptide treatment. The peptides inhibited growth of pneumococcal isolates of serotypes 3, 8, 9N, 12F and 19A, currently prevalent in Switzerland, and caused no detectable toxic effect to primary human airway epithelial cells. We conclude that pneumococci take up K. pneumoniae peptides from the environment via binding and transport through the Ami permease. This changes gene expression resulting in altered phenotypes, particularly reduced growth.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Cillóniz C, Garcia-Vidal C, Ceccato A, Torres A. Antimicrobial Resistance Among Streptococcus pneumoniae. In: Fong I, Shlaes D, Drlica K, editors. Antimicrobial Resistance in the 21st Century. Springer; 2018.

