Targeting ATP2B1 impairs PI3K/Akt/FOXO signaling and reduces SARS-COV-2 infection and replication
- PMID: 38816514
- PMCID: PMC11239940
- DOI: 10.1038/s44319-024-00164-z
Targeting ATP2B1 impairs PI3K/Akt/FOXO signaling and reduces SARS-COV-2 infection and replication
Abstract
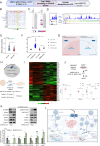
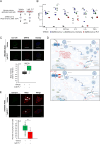
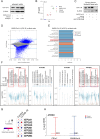
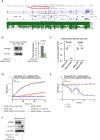
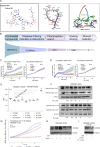
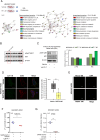
ATP2B1 is a known regulator of calcium (Ca2+) cellular export and homeostasis. Diminished levels of intracellular Ca2+ content have been suggested to impair SARS-CoV-2 replication. Here, we demonstrate that a nontoxic caloxin-derivative compound (PI-7) reduces intracellular Ca2+ levels and impairs SARS-CoV-2 infection. Furthermore, a rare homozygous intronic variant of ATP2B1 is shown to be associated with the severity of COVID-19. The mechanism of action during SARS-CoV-2 infection involves the PI3K/Akt signaling pathway activation, inactivation of FOXO3 transcription factor function, and subsequent transcriptional inhibition of the membrane and reticulum Ca2+ pumps ATP2B1 and ATP2A1, respectively. The pharmacological action of compound PI-7 on sustaining both ATP2B1 and ATP2A1 expression reduces the intracellular cytoplasmic Ca2+ pool and thus negatively influences SARS-CoV-2 replication and propagation. As compound PI-7 lacks toxicity in vitro, its prophylactic use as a therapeutic agent against COVID-19 is envisioned here.
Keywords: ATP2B1; Ca2+; PI3K/Akt/FOXO; SARS-CoV-2; Transcription.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Acheampong KK, Schaff DL, Emert BL, Lake J, Reffsin S, Shea EK, Comar CE, Litzky LA, Khurram NA, Linn RL, et al. Subcellular detection of SARS-CoV-2 RNA in human tissue reveals distinct localization in alveolar type 2 pneumocytes and alveolar macrophages. mBio. 2022;13:e0375121. doi: 10.1128/mbio.03751-21. - DOI - PMC - PubMed
-
- Andolfo I, Russo R, Lasorsa VA, Cantalupo S, Rosato BE, Bonfiglio F, Frisso G, Abete P, Cassese GM, Servillo G, Esposito G, Gentile I, Piscopo C, Villani R, Fiorentino G, Cerino P, Buonerba C, Pierri B, Zollo M, Iolascon A, Capasso M (2021) Common variants at 21q22.3 locus influence MX1 and TMPRSS2 gene expression and susceptibility to severe COVID-19. iScience 24:102322 - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

