Vaccination induces broadly neutralizing antibody precursors to HIV gp41
- PMID: 38816615
- PMCID: PMC11147780
- DOI: 10.1038/s41590-024-01833-w
Vaccination induces broadly neutralizing antibody precursors to HIV gp41
Erratum in
-
Author Correction: Vaccination induces broadly neutralizing antibody precursors to HIV gp41.Nat Immunol. 2024 Jul;25(7):1307. doi: 10.1038/s41590-024-01891-0. Nat Immunol. 2024. PMID: 38877179 Free PMC article. No abstract available.
Abstract
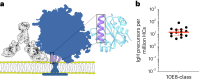
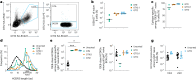
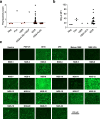
A key barrier to the development of vaccines that induce broadly neutralizing antibodies (bnAbs) against human immunodeficiency virus (HIV) and other viruses of high antigenic diversity is the design of priming immunogens that induce rare bnAb-precursor B cells. The high neutralization breadth of the HIV bnAb 10E8 makes elicitation of 10E8-class bnAbs desirable; however, the recessed epitope within gp41 makes envelope trimers poor priming immunogens and requires that 10E8-class bnAbs possess a long heavy chain complementarity determining region 3 (HCDR3) with a specific binding motif. We developed germline-targeting epitope scaffolds with affinity for 10E8-class precursors and engineered nanoparticles for multivalent display. Scaffolds exhibited epitope structural mimicry and bound bnAb-precursor human naive B cells in ex vivo screens, protein nanoparticles induced bnAb-precursor responses in stringent mouse models and rhesus macaques, and mRNA-encoded nanoparticles triggered similar responses in mice. Thus, germline-targeting epitope scaffold nanoparticles can elicit rare bnAb-precursor B cells with predefined binding specificities and HCDR3 features.
© 2024. The Author(s).
Conflict of interest statement
T.S., O.S., A.M.S., J.Z., F.S., D.W.K., G.G. and W.R.S. are named inventors on patent applications filed by Scripps and IAVI regarding 10E8-GT immunogens. S.H., S.P. and W.R.S. are employees of Moderna. G.G. is an employee of Generate Biomedicines. The other authors declare no competing interests.
Figures



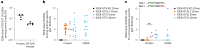










References
MeSH terms
Substances
Grants and funding
- P30 ES030283/ES/NIEHS NIH HHS/United States
- P51 OD011132/OD/NIH HHS/United States
- INV-002916/GATES/Gates Foundation/United States
- INV-009585/GATES/Gates Foundation/United States
- INV-008813/GATES/Gates Foundation/United States
- INV046626/Bill and Melinda Gates Foundation (Bill & Melinda Gates Foundation)
- P30 GM138396/GM/NIGMS NIH HHS/United States
- NV-007522/Bill and Melinda Gates Foundation (Bill & Melinda Gates Foundation)
- R01 AI147826/AI/NIAID NIH HHS/United States
- P30 AI036214/AI/NIAID NIH HHS/United States
- UM1 AI169662/AI/NIAID NIH HHS/United States
- UM1 AI144462/AI/NIAID NIH HHS/United States
- INV-007522/GATES/Gates Foundation/United States
- AI144462/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- U24 AI126683/AI/NIAID NIH HHS/United States
- U42 OD011023/OD/NIH HHS/United States
- AI147826/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

