Affinity gaps among B cells in germinal centers drive the selection of MPER precursors
- PMID: 38816616
- PMCID: PMC11147770
- DOI: 10.1038/s41590-024-01844-7
Affinity gaps among B cells in germinal centers drive the selection of MPER precursors
Abstract
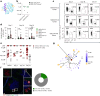
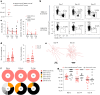
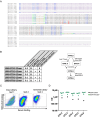
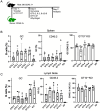
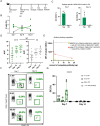
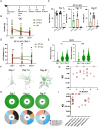
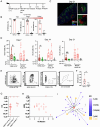
Current prophylactic human immunodeficiency virus 1 (HIV-1) vaccine research aims to elicit broadly neutralizing antibodies (bnAbs). Membrane-proximal external region (MPER)-targeting bnAbs, such as 10E8, provide exceptionally broad neutralization, but some are autoreactive. Here, we generated humanized B cell antigen receptor knock-in mouse models to test whether a series of germline-targeting immunogens could drive MPER-specific precursors toward bnAbs. We found that recruitment of 10E8 precursors to germinal centers (GCs) required a minimum affinity for germline-targeting immunogens, but the GC residency of MPER precursors was brief due to displacement by higher-affinity endogenous B cell competitors. Higher-affinity germline-targeting immunogens extended the GC residency of MPER precursors, but robust long-term GC residency and maturation were only observed for MPER-HuGL18, an MPER precursor clonotype able to close the affinity gap with endogenous B cell competitors in the GC. Thus, germline-targeting immunogens could induce MPER-targeting antibodies, and B cell residency in the GC may be regulated by a precursor-competitor affinity gap.
© 2024. The Author(s).
Conflict of interest statement
T.S., O.S. and W.R.S. are named inventors on patent applications filed by Scripps and IAVI regarding 10E8-GT immunogens and nanoparticles in this manuscript. F.D.B. has consultancy relationships with Adimab, Third Rock Ventures and
Figures











References
MeSH terms
Substances
Grants and funding
- OPP1147787/Bill and Melinda Gates Foundation (Bill & Melinda Gates Foundation)
- UM1 AI144462/AI/NIAID NIH HHS/United States
- INV009585/Bill and Melinda Gates Foundation (Bill & Melinda Gates Foundation)
- UM1 AI44462/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- INV-034657/GATES/Gates Foundation/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

