Alternative platelet differentiation pathways initiated by nonhierarchically related hematopoietic stem cells
- PMID: 38816617
- PMCID: PMC11147777
- DOI: 10.1038/s41590-024-01845-6
Alternative platelet differentiation pathways initiated by nonhierarchically related hematopoietic stem cells
Abstract
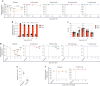
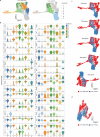
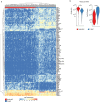
Rare multipotent stem cells replenish millions of blood cells per second through a time-consuming process, passing through multiple stages of increasingly lineage-restricted progenitors. Although insults to the blood-forming system highlight the need for more rapid blood replenishment from stem cells, established models of hematopoiesis implicate only one mandatory differentiation pathway for each blood cell lineage. Here, we establish a nonhierarchical relationship between distinct stem cells that replenish all blood cell lineages and stem cells that replenish almost exclusively platelets, a lineage essential for hemostasis and with important roles in both the innate and adaptive immune systems. These distinct stem cells use cellularly, molecularly and functionally separate pathways for the replenishment of molecularly distinct megakaryocyte-restricted progenitors: a slower steady-state multipotent pathway and a fast-track emergency-activated platelet-restricted pathway. These findings provide a framework for enhancing platelet replenishment in settings in which slow recovery of platelets remains a major clinical challenge.
© 2024. The Author(s).
Conflict of interest statement
M.H.-J., C.Z. and R.S. report holding shares in Xpress Genomics AB, and M.H.-J. and R.S. are inventors on the patent relating to Smart-seq3 that is licensed to Takara Bio USA. The other authors declare no competing interests.
Figures
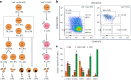


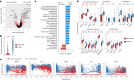
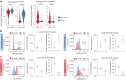
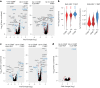









References
MeSH terms
Grants and funding
- MC_UU_12009/5/RCUK | Medical Research Council (MRC)
- 224132/Radiumhemmets Forskningsfonder (Cancer Research Foundations of Radiumhemmet)
- 26988/CRUK_/Cancer Research UK/United Kingdom
- 538-2013-8995/Vetenskapsrådet (Swedish Research Council)
- KAW 2015.0195/Knut och Alice Wallenbergs Stiftelse (Knut and Alice Wallenberg Foundation)
LinkOut - more resources
Full Text Sources
Medical

