Targeting a lineage-specific PI3Kɣ-Akt signaling module in acute myeloid leukemia using a heterobifunctional degrader molecule
- PMID: 38816660
- PMCID: PMC11778622
- DOI: 10.1038/s43018-024-00782-5
Targeting a lineage-specific PI3Kɣ-Akt signaling module in acute myeloid leukemia using a heterobifunctional degrader molecule
Abstract
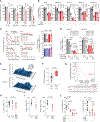
Dose-limiting toxicity poses a major limitation to the clinical utility of targeted cancer therapies, often arising from target engagement in nonmalignant tissues. This obstacle can be minimized by targeting cancer dependencies driven by proteins with tissue-restricted and/or tumor-restricted expression. In line with another recent report, we show here that, in acute myeloid leukemia (AML), suppression of the myeloid-restricted PIK3CG/p110γ-PIK3R5/p101 axis inhibits protein kinase B/Akt signaling and compromises AML cell fitness. Furthermore, silencing the genes encoding PIK3CG/p110γ or PIK3R5/p101 sensitizes AML cells to established AML therapies. Importantly, we find that existing small-molecule inhibitors against PIK3CG are insufficient to achieve a sustained long-term antileukemic effect. To address this concern, we developed a proteolysis-targeting chimera (PROTAC) heterobifunctional molecule that specifically degrades PIK3CG and potently suppresses AML progression alone and in combination with venetoclax in human AML cell lines, primary samples from patients with AML and syngeneic mouse models.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
The Krogan Laboratory has received research support from Vir Biotechnology, F. Hoffmann-La Roche, and Rezo Therapeutics. N.J.K. has financially compensated consulting agreements with Maze Therapeutics. N.J.K. is the President and is on the Board of Directors of Rezo Therapeutics, and he is a shareholder in Tenaya Therapeutics, Maze Therapeutics, Rezo Therapeutics, and Interline Therapeutics. K.C.W. is a co-founder, consultant, and equity holder at Tavros Therapeutics and Celldom, is a consultant and equity holder at Simple Therapeutics and Decrypt Biomedicine, and has performed consulting work for Guidepoint Global, Bantam Pharmaceuticals, and Apple Tree Partners. The remaining authors declare no competing interests.
Figures





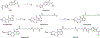








References
References to the Methods section
-
- Su A. et al. The Folate Cycle Enzyme MTHFR Is a Critical Regulator of Cell Response to MYC-Targeting Therapies. Cancer discovery 10, 1894–1911, doi:10.1158/2159-8290.CD-19-0970 (2020). - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 101088563/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- P41 GM103504/GM/NIGMS NIH HHS/United States
- 5U54CA274502-02/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- U54 CA274502/CA/NCI NIH HHS/United States
- PLBIO20-074/Institut National Du Cancer (French National Cancer Institute)
- 758848/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- R01CA266389/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- SIRIC InsiTu program (INCa-DGOS-INSERM-ITMO Cancer_18008)/Institut National Du Cancer (French National Cancer Institute)
- R01 CA266389/CA/NCI NIH HHS/United States
- CCA INSERM Bettencourt/Fondation Bettencourt Schueller (Bettencourt Schueller Foundation)
- PLBIO20-246/Institut National Du Cancer (French National Cancer Institute)
- 5U54CA274502-02/U.S. Department of Health & Human Services | NIH | U.S. National Library of Medicine (NLM)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

