MFPINC: prediction of plant ncRNAs based on multi-source feature fusion
- PMID: 38816689
- PMCID: PMC11137975
- DOI: 10.1186/s12864-024-10439-3
MFPINC: prediction of plant ncRNAs based on multi-source feature fusion
Abstract
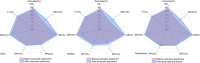
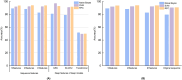
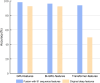
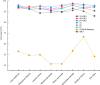
Non-coding RNAs (ncRNAs) are recognized as pivotal players in the regulation of essential physiological processes such as nutrient homeostasis, development, and stress responses in plants. Common methods for predicting ncRNAs are susceptible to significant effects of experimental conditions and computational methods, resulting in the need for significant investment of time and resources. Therefore, we constructed an ncRNA predictor(MFPINC), to predict potential ncRNA in plants which is based on the PINC tool proposed by our previous studies. Specifically, sequence features were carefully refined using variance thresholding and F-test methods, while deep features were extracted and feature fusion were performed by applying the GRU model. The comprehensive evaluation of multiple standard datasets shows that MFPINC not only achieves more comprehensive and accurate identification of gene sequences, but also significantly improves the expressive and generalization performance of the model, and MFPINC significantly outperforms the existing competing methods in ncRNA identification. In addition, it is worth mentioning that our tool can also be found on Github ( https://github.com/Zhenj-Nie/MFPINC ) the data and source code can also be downloaded for free.
Keywords: Fusion of deep feature and sequence feature; Plants; ncRNA prediction.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests
Figures








Similar articles
-
PINC: A Tool for Non-Coding RNA Identification in Plants Based on an Automated Machine Learning Framework.Int J Mol Sci. 2022 Oct 5;23(19):11825. doi: 10.3390/ijms231911825. Int J Mol Sci. 2022. PMID: 36233123 Free PMC article.
-
MFPred: prediction of ncRNA families based on multi-feature fusion.Brief Bioinform. 2023 Sep 20;24(5):bbad303. doi: 10.1093/bib/bbad303. Brief Bioinform. 2023. PMID: 37615358
-
Improving ncRNA family prediction using multi-modal contrastive learning of sequence and structure.Bioinformatics. 2024 Nov 1;40(11):btae640. doi: 10.1093/bioinformatics/btae640. Bioinformatics. 2024. PMID: 39460948 Free PMC article.
-
Biogenesis, Functions, Interactions, and Resources of Non-Coding RNAs in Plants.Int J Mol Sci. 2022 Mar 28;23(7):3695. doi: 10.3390/ijms23073695. Int J Mol Sci. 2022. PMID: 35409060 Free PMC article. Review.
-
Advances in Computational Methodologies for Classification and Sub-Cellular Locality Prediction of Non-Coding RNAs.Int J Mol Sci. 2021 Aug 13;22(16):8719. doi: 10.3390/ijms22168719. Int J Mol Sci. 2021. PMID: 34445436 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

