Immune cell infiltration and inflammatory landscape in primary brain tumours
- PMID: 38816839
- PMCID: PMC11140972
- DOI: 10.1186/s12967-024-05309-1
Immune cell infiltration and inflammatory landscape in primary brain tumours
Abstract
Background: Primary malignant brain tumours are more than one-third of all brain tumours and despite the molecular investigation to identify cancer driver mutations, the current therapeutic options available are challenging due to high intratumour heterogeneity. In addition, an immunosuppressive and inflammatory tumour microenvironment strengthens cancer progression. Therefore, we defined an immune and inflammatory profiling of meningioma and glial tumours to elucidate the role of the immune infiltration in these cancer types.
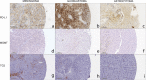
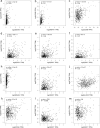
Methods: Using tissue microarrays of 158 brain tumour samples, we assessed CD3, CD4, CD8, CD20, CD138, Granzyme B (GzmB), 5-Lipoxygenase (5-LOX), Programmed Death-Ligand 1 (PD-L1), O-6-Methylguanine-DNA Methyltransferase (MGMT) and Transglutaminase 2 (TG2) expression by immunohistochemistry (IHC). IHC results were correlated using a Spearman correlation matrix. Transcript expression, correlation, and overall survival (OS) analyses were evaluated using public datasets available on GEPIA2 in Glioblastoma (GBM) and Lower Grade Glioma (LGG) cohorts.
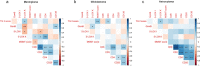
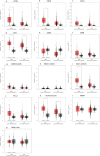
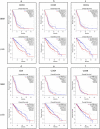
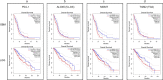
Results: Seven out of ten markers showed a significantly different IHC expression in at least one of the evaluated cohorts whereas CD3, CD4 and 5-LOX were differentially expressed between GBMs and astrocytomas. Correlation matrix analysis revealed that 5-LOX and GzmB expression were associated in both meningiomas and GBMs, whereas 5-LOX expression was significantly and positively correlated to TG2 in both meningioma and astrocytoma cohorts. These findings were confirmed with the correlation analysis of TCGA-GBM and LGG datasets. Profiling of mRNA levels indicated a significant increase in CD3 (CD3D, CD3E), and CD138 (SDC1) expression in GBM compared to control tissues. CD4 and 5-LOX (ALOX5) mRNA levels were significantly more expressed in tumour samples than in normal tissues in both GBM and LGG. In GBM cohort, GzmB (GZMB), SDC1 and MGMT gene expression predicted a poor overall survival (OS). Moreover, in LGG cohort, an increased expression of CD3 (CD3D, CD3E, CD3G), CD8 (CD8A), GZMB, CD20 (MS4A1), SDC1, PD-L1, ALOX5, and TG2 (TGM2) genes was associated with worse OS.
Conclusions: Our data have revealed that there is a positive and significant correlation between the expression of 5-LOX and GzmB, both at RNA and protein level. Further evaluation is needed to understand the interplay of 5-LOX and immune infiltration in glioma progression.
Keywords: 5-Lypoxygenase; Astrocytoma; Glioblastoma; Immune cell infiltration; Inflammation; Meningioma.
© 2024. The Author(s).
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures









References
-
- Newton HB. Chapter 1—overview of brain tumor epidemiology. In: Newton HB, editor. Handbook of neuro-oncology neuroimaging (Second Edition) San Diego: Academic Press; 2016. pp. 3–8.
-
- Reynoso-Noverón N, Mohar-Betancourt A, Ortiz-Rafael J. Epidemiology of brain tumors. In: Monroy-Sosa A, Chakravarthi SS, de la Garza-Salazar JG, Meneses Garcia A, Kassam AB, editors. Principles of neuro-oncology: brain & skull base. Cham: Springer International Publishing; 2021. pp. 15–25.
Publication types
MeSH terms
Grants and funding
- Research/Ministero dell'Istruzione, dell'Università e della Ricerca
- Innovation 2014-2020 - AIM Attraction/Ministero dell'Istruzione, dell'Università e della Ricerca
- International Mobility/Ministero dell'Istruzione, dell'Università e della Ricerca
- Epi-MET-Funzionalizzazione delle aberrazioni (epi)genomiche nei tumori metastatici"/Ministero dello Sviluppo Economico
- grant number: F/310034/01-03/X56./Ministero dello Sviluppo Economico
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

