Advancing skeletal health and disease research with single-cell RNA sequencing
- PMID: 38816888
- PMCID: PMC11138034
- DOI: 10.1186/s40779-024-00538-3
Advancing skeletal health and disease research with single-cell RNA sequencing
Abstract
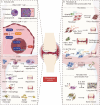
Orthopedic conditions have emerged as global health concerns, impacting approximately 1.7 billion individuals worldwide. However, the limited understanding of the underlying pathological processes at the cellular and molecular level has hindered the development of comprehensive treatment options for these disorders. The advent of single-cell RNA sequencing (scRNA-seq) technology has revolutionized biomedical research by enabling detailed examination of cellular and molecular diversity. Nevertheless, investigating mechanisms at the single-cell level in highly mineralized skeletal tissue poses technical challenges. In this comprehensive review, we present a streamlined approach to obtaining high-quality single cells from skeletal tissue and provide an overview of existing scRNA-seq technologies employed in skeletal studies along with practical bioinformatic analysis pipelines. By utilizing these methodologies, crucial insights into the developmental dynamics, maintenance of homeostasis, and pathological processes involved in spine, joint, bone, muscle, and tendon disorders have been uncovered. Specifically focusing on the joint diseases of degenerative disc disease, osteoarthritis, and rheumatoid arthritis using scRNA-seq has provided novel insights and a more nuanced comprehension. These findings have paved the way for discovering novel therapeutic targets that offer potential benefits to patients suffering from diverse skeletal disorders.
Keywords: Bioinformatic analysis; Cellular heterogeneity; Musculoskeletal system; Single cell suspension; Single-cell RNA sequencing (scRNA-seq); Skeletal disorders.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
Publication types
MeSH terms
Grants and funding
- 2022YFA1103202/National Key Research and Development Program of China
- 82272507/National Natural Science Foundation of China
- 32270887/National Natural Science Foundation of China
- 32200654/National Natural Science Foundation of China
- CSTB2023NSCQ-ZDJO008/Natural Science Foundation of Chongqing
- BX20220397/Postdoctoral Innovative Talent Support Program
- SFLKF202201/Independent Research Project of State Key Laboratory of Trauma and Chemical Poisoning
- 2021-XZYG-B10/General Hospital of Western Theater Command Research Project
- 14113723/University Grants Committee, Research Grants Council of Hong Kong, China
- N_CUHK472/22/University Grants Committee, Research Grants Council of Hong Kong, China
- C7030-18G/University Grants Committee, Research Grants Council of Hong Kong, China
- T13-402/17-N/University Grants Committee, Research Grants Council of Hong Kong, China
- AoE/M-402/20/University Grants Committee, Research Grants Council of Hong Kong, China
LinkOut - more resources
Full Text Sources

