Brassinosteroid biosynthesis and signaling: Conserved and diversified functions of core genes across multiple plant species
- PMID: 38816993
- PMCID: PMC11412936
- DOI: 10.1016/j.xplc.2024.100982
Brassinosteroid biosynthesis and signaling: Conserved and diversified functions of core genes across multiple plant species
Abstract
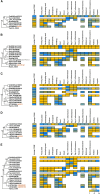
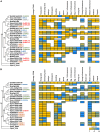
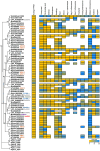
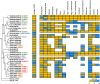
Brassinosteroids (BRs) are important regulators that control myriad aspects of plant growth and development, including biotic and abiotic stress responses, such that modulating BR homeostasis and signaling presents abundant opportunities for plant breeding and crop improvement. Enzymes and other proteins involved in the biosynthesis and signaling of BRs are well understood from molecular genetics and phenotypic analysis in Arabidopsis thaliana; however, knowledge of the molecular functions of these genes in other plant species, especially cereal crop plants, is minimal. In this manuscript, we comprehensively review functional studies of BR genes in Arabidopsis, maize, rice, Setaria, Brachypodium, and soybean to identify conserved and diversified functions across plant species and to highlight cases for which additional research is in order. We performed phylogenetic analysis of gene families involved in the biosynthesis and signaling of BRs and re-analyzed publicly available transcriptomic data. Gene trees coupled with expression data provide a valuable guide to supplement future research on BRs in these important crop species, enabling researchers to identify gene-editing targets for BR-related functional studies.
Keywords: Arabidopsis; Brachypodium; Setaria; brassinosteroids; maize; rice; soybean.
Published by Elsevier Inc.
Figures



Similar articles
-
Identification of brassinosteroid genes in Brachypodium distachyon.BMC Plant Biol. 2017 Jan 6;17(1):5. doi: 10.1186/s12870-016-0965-3. BMC Plant Biol. 2017. PMID: 28061864 Free PMC article.
-
GmBZL3 acts as a major BR signaling regulator through crosstalk with multiple pathways in Glycine max.BMC Plant Biol. 2019 Feb 22;19(1):86. doi: 10.1186/s12870-019-1677-2. BMC Plant Biol. 2019. PMID: 30795735 Free PMC article.
-
Down-regulation of BdBRI1, a putative brassinosteroid receptor gene produces a dwarf phenotype with enhanced drought tolerance in Brachypodium distachyon.Plant Sci. 2015 May;234:163-73. doi: 10.1016/j.plantsci.2015.02.015. Epub 2015 Feb 27. Plant Sci. 2015. PMID: 25804819
-
Benefits of brassinosteroid crosstalk.Trends Plant Sci. 2012 Oct;17(10):594-605. doi: 10.1016/j.tplants.2012.05.012. Epub 2012 Jun 26. Trends Plant Sci. 2012. PMID: 22738940 Review.
-
Auxin EvoDevo: Conservation and Diversification of Genes Regulating Auxin Biosynthesis, Transport, and Signaling.Mol Plant. 2019 Mar 4;12(3):298-320. doi: 10.1016/j.molp.2018.12.012. Epub 2018 Dec 25. Mol Plant. 2019. PMID: 30590136 Review.
Cited by
-
A Comparative Analysis of the Effect of 24-Epibrassinolide on the Tolerance of Wheat Cultivars with Different Drought Adaptation Strategies Under Water Deficit Conditions.Plants (Basel). 2025 Mar 10;14(6):869. doi: 10.3390/plants14060869. Plants (Basel). 2025. PMID: 40265790 Free PMC article.
-
Time-Course Transcriptomics Analysis Reveals Molecular Mechanisms of Salt-Tolerant and Salt-Sensitive Cotton Cultivars in Response to Salt Stress.Int J Mol Sci. 2025 Jan 2;26(1):329. doi: 10.3390/ijms26010329. Int J Mol Sci. 2025. PMID: 39796184 Free PMC article.
-
Preharvest Application of Exogenous 2,4-Epibrassinolide and Melatonin Enhances the Maturity and Flue-Cured Quality of Tobacco Leaves.Plants (Basel). 2024 Nov 21;13(23):3266. doi: 10.3390/plants13233266. Plants (Basel). 2024. PMID: 39683059 Free PMC article.
-
STay tunEd: mutational analysis of the HvSTE1 gene in barley provides insight into the balance between semi-dwarfism and maintenance of grain size in brassinosteroid biosynthesis-dependent manner.Front Plant Sci. 2025 May 6;16:1571368. doi: 10.3389/fpls.2025.1571368. eCollection 2025. Front Plant Sci. 2025. PMID: 40395283 Free PMC article.
-
The Evolution of Plant Hormones: From Metabolic Byproducts to Regulatory Hubs.Int J Mol Sci. 2025 Jul 25;26(15):7190. doi: 10.3390/ijms26157190. Int J Mol Sci. 2025. PMID: 40806323 Free PMC article. Review.
References
-
- Albrecht C., Boutrot F., Segonzac C., Schwessinger B., Gimenez-Ibanez S., Chinchilla D., Rathjen J.P., de Vries S.C., Zipfel C. Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl. Acad. Sci. USA. 2012;109:303–308. doi: 10.1073/pnas.1109921108. - DOI - PMC - PubMed
-
- Babiychuk E., Bouvier-Navé P., Compagnon V., Suzuki M., Muranaka T., Van Montagu M., Kushnir S., Schaller H. Allelic mutant series reveal distinct functions for Arabidopsis cycloartenol synthase 1 in cell viability and plastid biogenesis. Proc. Natl. Acad. Sci. USA. 2008;105:3163–3168. doi: 10.1073/pnas.0712190105. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

