Developing a Predictive Model for Metastatic Potential in Pancreatic Neuroendocrine Tumor
- PMID: 38817124
- PMCID: PMC11651689
- DOI: 10.1210/clinem/dgae380
Developing a Predictive Model for Metastatic Potential in Pancreatic Neuroendocrine Tumor
Abstract
Context: Pancreatic neuroendocrine tumors (PNETs) exhibit a wide range of behavior from localized disease to aggressive metastasis. A comprehensive transcriptomic profile capable of differentiating between these phenotypes remains elusive.
Objective: Use machine learning to develop predictive models of PNET metastatic potential dependent upon transcriptomic signature.
Methods: RNA-sequencing data were analyzed from 95 surgically resected primary PNETs in an international cohort. Two cohorts were generated with equally balanced metastatic PNET composition. Machine learning was used to create predictive models distinguishing between localized and metastatic tumors. Models were validated on an independent cohort of 29 formalin-fixed, paraffin-embedded samples using NanoString nCounter®, a clinically available mRNA quantification platform.
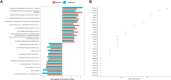
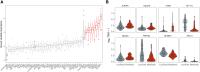
Results: Gene expression analysis identified concordant differentially expressed genes between the 2 cohorts. Gene set enrichment analysis identified additional genes that contributed to enriched biologic pathways in metastatic PNETs. Expression values for these genes were combined with an additional 7 genes known to contribute to PNET oncogenesis and prognosis, including ARX and PDX1. Eight specific genes (AURKA, CDCA8, CPB2, MYT1L, NDC80, PAPPA2, SFMBT1, ZPLD1) were identified as sufficient to classify the metastatic status with high sensitivity (87.5-93.8%) and specificity (78.1-96.9%). These models remained predictive of the metastatic phenotype using NanoString nCounter® on the independent validation cohort, achieving a median area under the receiving operating characteristic curve of 0.886.
Conclusion: We identified and validated an 8-gene panel predictive of the metastatic phenotype in PNETs, which can be detected using the clinically available NanoString nCounter® system. This panel should be studied prospectively to determine its utility in guiding operative vs nonoperative management.
Keywords: PNET; machine learning; neuroendocrine tumor; pancreatic neuroendocrine tumor.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Endocrine Society. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com. See the journal About page for additional terms.
Figures



References
-
- Yao JC, Hassan M, Phan A, et al. One hundred years after “carcinoid”: epidemiology of and prognostic factors for neuroendocrine tumors in 35,825 cases in the United States. J Clin Oncol. 2008;26(18):3063‐3072. - PubMed
-
- Lloyd R, Osamura R, Klöppel G, Rosai J. WHO Classification of Tumours of Endocrine Organs. IARC Press; 2017.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

