Whole-genome sequencing uncovers two loci for coronary artery calcification and identifies ARSE as a regulator of vascular calcification
- PMID: 38817323
- PMCID: PMC11138106
- DOI: 10.1038/s44161-023-00375-y
Whole-genome sequencing uncovers two loci for coronary artery calcification and identifies ARSE as a regulator of vascular calcification
Abstract
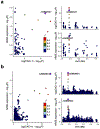
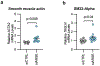
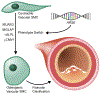
Coronary artery calcification (CAC) is a measure of atherosclerosis and a well-established predictor of coronary artery disease (CAD) events. Here we describe a genome-wide association study (GWAS) of CAC in 22,400 participants from multiple ancestral groups. We confirmed associations with four known loci and identified two additional loci associated with CAC (ARSE and MMP16), with evidence of significant associations in replication analyses for both novel loci. Functional assays of ARSE and MMP16 in human vascular smooth muscle cells (VSMCs) demonstrate that ARSE is a promoter of VSMC calcification and VSMC phenotype switching from a contractile to a calcifying or osteogenic phenotype. Furthermore, we show that the association of variants near ARSE with reduced CAC is likely explained by reduced ARSE expression with the G allele of enhancer variant rs5982944. Our study highlights ARSE as an important contributor to atherosclerotic vascular calcification, and a potential drug target for vascular calcific disease.
Conflict of interest statement
Bruce M. Psaty serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. Leslie S. Emery is now an employee of Celgene/Bristol Myers Squibb. Celgene/Bristol Myers Squibb had no role in the funding, design, conduct, or interpretation of this study. Nathan O. Stitziel has received research funding from Regeneron Pharmaceuticals, unrelated to this work. Karine A. Viaud-Martinez is an employee at Illumina Inc. Ryan W. Kim is an employee at Psomagen Inc. Roxona Mehran reports institutional research grants from Abbott Laboratories, Abiomed, Applied Therapeutics, AstraZeneca, Bayer, Beth Israel Deaconess, Bristol-Myers Squibb, CERC, Chiesi, Concept Medical, CSL Behring, DSI, Medtronic, Novartis Pharmaceuticals, OrbusNeich; consultant fees from Abbott Laboratories, Boston Scientific, CardiaWave, Chiesi, Janssen Scientific Affairs, Medscape/WebMD, Medtelligence (Janssen Scientific Affairs), Roivant Sciences, Sanofi, Siemens Medical Solutions; consultant fees paid to the institution from Abbott Laboratories, Bristol-Myers Squibb; advisory board, funding paid to the institution from Spectranetics/Philips/Volcano Corp; consultant (spouse) from Abiomed, The Medicines Company, Merck; Equity <1% from Claret Medical, Elixir Medical; DSMB Membership fees paid to the institution from Watermark Research Partners; consulting (no fee) from Idorsia Pharmaceuticals Ltd., Regeneron Pharmaceuticals; Associate Editor for ACC, AMA. Rajeev Malhotra is a consultant for MyoKardia (now owned by BMS), Epizon Pharma, Renovacor, and Third Pole, a co-founder of Patch and Angea Biotherapeutics, and has received research funding from Angea Biotherapeutics, Bayer Pharmaceuticals, and Amgen. Adrienne M. Stilp receives funding from Seven Bridges Genomics to develop tools for the NHLBI BioData Catalyst consortium. Pradeep Natarajan reports grants from Amgen, Apple, Boston Scientific, AstraZeneca, Allelica, Novartis, and Genentech, consulting income from GV, Blackstone Life Sciences, Foresite Labs, Apple, AstraZeneca, Allelica, Novartis, HeartFlow, and Genentech, is a scientific advisor to Esperion Therapeutics, Preciseli, and TenSixteen Bio, is a scientific co-founder of TenSixteen Bio, and spousal employment at Vertex, all unrelated to the present work. Clint L. Miller received a research grant from AstraZeneca for an unrelated project. The remaining authors declare no competing interests.
Figures

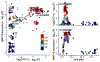

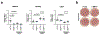



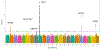

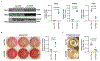
References
-
- Kavousi M et al. Evaluation of newer risk markers for coronary heart disease risk classification: a cohort study. Ann Intern Med 156, 438–44 (2012). - PubMed
METHODS-ONLY REFERENCES
-
- Agatston AS et al. Quantification of coronary artery calcium using ultrafast computed tomography. J Am Coll Cardiol 15, 827–32 (1990). - PubMed
-
- Carr JJ et al. Calcified coronary artery plaque measurement with cardiac CT in population-based studies: standardized protocol of Multi-Ethnic Study of Atherosclerosis (MESA) and Coronary Artery Risk Development in Young Adults (CARDIA) study. Radiology 234, 35–43 (2005). - PubMed
Grants and funding
- R01 HL120393/HL/NHLBI NIH HHS/United States
- R01 EB015611/EB/NIBIB NIH HHS/United States
- U01 HL120393/HL/NHLBI NIH HHS/United States
- R01 HL162928/HL/NHLBI NIH HHS/United States
- R01 HL146860/HL/NHLBI NIH HHS/United States
- R01 HL164577/HL/NHLBI NIH HHS/United States
- R01 HL148239/HL/NHLBI NIH HHS/United States
- U01 HL137162/HL/NHLBI NIH HHS/United States
- R01 HL117626/HL/NHLBI NIH HHS/United States
- R01 HL142809/HL/NHLBI NIH HHS/United States
- R01 HL159514/HL/NHLBI NIH HHS/United States
- R01 HL142711/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
