Single-cell Raman and functional gene analyses reveal microbial P solubilization in agriculture waste-modified soils
- PMID: 38817623
- PMCID: PMC10989763
- DOI: 10.1002/mlf2.12053
Single-cell Raman and functional gene analyses reveal microbial P solubilization in agriculture waste-modified soils
Abstract
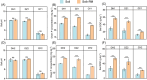
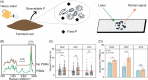
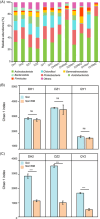
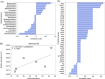
Application of agricultural waste such as rapeseed meal (RM) is regarded as a sustainable way to improve soil phosphorus (P) availability by direct nutrient supply and stimulation of native phosphate-solubilizing microorganisms (PSMs) in soils. However, exploration of the in situ microbial P solubilizing function in soils remains a challenge. Here, by applying both phenotype-based single-cell Raman with D2O labeling (Raman-D2O) and genotype-based high-throughput chips targeting carbon, nitrogen and P (CNP) functional genes, the effect of RM application on microbial P solubilization in three typical farmland soils was investigated. The abundances of PSMs increased in two alkaline soils after RM application identified by single-cell Raman D2O. RM application reduced the diversity of bacterial communities and increased the abundance of a few bacteria with reported P solubilization function. Genotypic analysis indicated that RM addition generally increased the relative abundance of CNP functional genes. A correlation analysis of the abundance of active PSMs with the abundance of soil microbes or functional genes was carried out to decipher the linkage between the phenotype and genotype of PSMs. Myxococcota and C degradation genes were found to potentially contribute to the enhanced microbial P release following RM application. This work provides important new insights into the in situ function of soil PSMs. It will lead to better harnessing of agricultural waste to mobilize soil legacy P and mitigate the P crisis.
Keywords: CNP functional genes; D2O isotope labeling; phosphate‐solubilizing microorganisms; single‐cell Raman.
© 2023 The Authors. mLife published by John Wiley & Sons Australia, Ltd. on behalf of Institute of Microbiology, Chinese Academy of Sciences.
Conflict of interest statement
The authors declare no conflict of interests.
Figures


Similar articles
-
D2O-Isotope-Labeling Approach to Probing Phosphate-Solubilizing Bacteria in Complex Soil Communities by Single-Cell Raman Spectroscopy.Anal Chem. 2019 Feb 5;91(3):2239-2246. doi: 10.1021/acs.analchem.8b04820. Epub 2019 Jan 16. Anal Chem. 2019. PMID: 30608659
-
Genome-Resolved Metagenomics Reveals Distinct Phosphorus Acquisition Strategies between Soil Microbiomes.mSystems. 2022 Feb 22;7(1):e0110721. doi: 10.1128/msystems.01107-21. Epub 2022 Jan 11. mSystems. 2022. PMID: 35014868 Free PMC article.
-
Prospects of phosphate solubilizing microorganisms in sustainable agriculture.World J Microbiol Biotechnol. 2024 Aug 6;40(10):291. doi: 10.1007/s11274-024-04086-9. World J Microbiol Biotechnol. 2024. PMID: 39105959 Review.
-
Distribution of Culturable Phosphate-Solubilizing Bacteria in Soil Aggregates and Their Potential for Phosphorus Acquisition.Microbiol Spectr. 2022 Jun 29;10(3):e0029022. doi: 10.1128/spectrum.00290-22. Epub 2022 May 10. Microbiol Spectr. 2022. PMID: 35536021 Free PMC article.
-
Soil phosphorus transformation and plant uptake driven by phosphate-solubilizing microorganisms.Front Microbiol. 2024 Mar 27;15:1383813. doi: 10.3389/fmicb.2024.1383813. eCollection 2024. Front Microbiol. 2024. PMID: 38601943 Free PMC article. Review.
Cited by
-
Single-cell exploration of active phosphate-solubilizing bacteria across diverse soil matrices for sustainable phosphorus management.Nat Food. 2024 Aug;5(8):673-683. doi: 10.1038/s43016-024-01024-8. Epub 2024 Aug 5. Nat Food. 2024. PMID: 39103543
References
-
- Yang X, Post WM, Thornton PE, Jain A. The distribution of soil phosphorus for global biogeochemical modeling. Biogeosciences. 2013;10:2525–37.
-
- Sarkhot DV, Ghezzehei TA, Berhe AA. Effectiveness of biochar for sorption of ammonium and phosphate from dairy effluent. J Environ Qual. 2013;42:1545–54. - PubMed
-
- Zhu J, Li M, Whelan M. Phosphorus activators contribute to legacy phosphorus availability in agricultural soils: a review. Sci Total Environ. 2018;612:522–37. - PubMed
-
- Peñuelas J, Poulter B, Sardans J, Ciais P, Van Der Velde M, Bopp L, et al. Human‐induced nitrogen–phosphorus imbalances alter natural and managed ecosystems across the globe. Nat Commun. 2013;4:2934. - PubMed
LinkOut - more resources
Full Text Sources
