The secretory Candida effector Sce1 licenses fungal virulence by masking the immunogenic β-1,3-glucan and promoting apoptosis of the host cells
- PMID: 38817625
- PMCID: PMC10989805
- DOI: 10.1002/mlf2.12066
The secretory Candida effector Sce1 licenses fungal virulence by masking the immunogenic β-1,3-glucan and promoting apoptosis of the host cells
Abstract
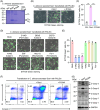
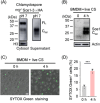
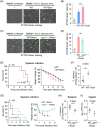
Candida albicans deploys a variety of mechanisms such as morphological switch and elicitor release to promote virulence. However, the intricate interactions between the fungus and the host remain poorly understood, and a comprehensive inventory of fungal virulence factors has yet to be established. In this study, we identified a C. albicans secretory effector protein Sce1, whose induction and secretion are associated with vagina-simulative conditions and chlamydospore formation. Sequence alignment showed that Sce1 belongs to a Pir family in C. albicans, which is conserved across several fungi and primarily characterized as a β-glucan binding protein in the Saccharomyces cerevisiae. Mechanically, Sce1 is primarily localized to the cell wall in a cleaved form as an alkali-labile β-1,3-glucan binding protein and plays a role in masking β-glucan in acidic environments and chlamydospores, a feature that might underline C. albicans' ability to evade host immunity. Further, a cleaved short form of Sce1 protein could be released into extracellular compartments and presented in bone marrow-derived macrophages infected with chlamydospores. This cleaved short form of Sce1 also demonstrated a unique ability to trigger the caspases-8/9-dependent apoptosis in various host cells. Correspondingly, genetic deletion of SCE1 led to dampened vaginal colonization of C. albicans and diminished fungal virulence during systemic infection. The discovery of Sce1 as a versatile virulence effector that executes at various compartments sheds light on the fungus-host interactions and C. albicans pathogenesis.
Keywords: Candida albicans; apoptosis; effector; immune evasion; β‐glucan.
© 2023 The Authors. mLife published by John Wiley & Sons Australia, Ltd. on behalf of Institute of Microbiology, Chinese Academy of Sciences.
Conflict of interest statement
The authors declare no conflict of interests.
Figures






Similar articles
-
Hypoxia Promotes Immune Evasion by Triggering β-Glucan Masking on the Candida albicans Cell Surface via Mitochondrial and cAMP-Protein Kinase A Signaling.mBio. 2018 Nov 6;9(6):e01318-18. doi: 10.1128/mBio.01318-18. mBio. 2018. PMID: 30401773 Free PMC article.
-
Epitope Shaving Promotes Fungal Immune Evasion.mBio. 2020 Jul 7;11(4):e00984-20. doi: 10.1128/mBio.00984-20. mBio. 2020. PMID: 32636248 Free PMC article.
-
Lrg1 Regulates β (1,3)-Glucan Masking in Candida albicans through the Cek1 MAP Kinase Pathway.mBio. 2019 Sep 17;10(5):e01767-19. doi: 10.1128/mBio.01767-19. mBio. 2019. PMID: 31530671 Free PMC article.
-
Revisiting the Vital Drivers and Mechanisms of β-Glucan Masking in Human Fungal Pathogen, Candida albicans.Pathogens. 2021 Jul 27;10(8):942. doi: 10.3390/pathogens10080942. Pathogens. 2021. PMID: 34451406 Free PMC article. Review.
-
Cell wall associated proteins involved in filamentation with impact on the virulence of Candida albicans.Microbiol Res. 2022 May;258:126996. doi: 10.1016/j.micres.2022.126996. Epub 2022 Feb 22. Microbiol Res. 2022. PMID: 35247799 Review.
References
-
- Pekmezovic M, Hovhannisyan H, Gresnigt MS, Iracane E, Oliveira‐Pacheco J, Siscar‐Lewin S, et al. Candida pathogens induce protective mitochondria‐associated type I interferon signalling and a damage‐driven response in vaginal epithelial cells. Nat Microbiol. 2021;6:643–57. - PubMed
LinkOut - more resources
Full Text Sources
