A bacterial symbiont in the gill of the marine scallop Argopecten irradians irradians metabolizes dimethylsulfoniopropionate
- PMID: 38817626
- PMCID: PMC10989825
- DOI: 10.1002/mlf2.12072
A bacterial symbiont in the gill of the marine scallop Argopecten irradians irradians metabolizes dimethylsulfoniopropionate
Abstract
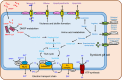
Microbial lysis of dimethylsulfoniopropionate (DMSP) is a key step in marine organic sulfur cycling and has been recently demonstrated to play an important role in mediating interactions between bacteria, algae, and zooplankton. To date, microbes that have been found to lyse DMSP are largely confined to free-living and surface-attached bacteria. In this study, we report for the first time that a symbiont (termed "Rhodobiaceae bacterium HWgs001") in the gill of the marine scallop Argopecten irradians irradians can lyse and metabolize DMSP. Analysis of 16S rRNA gene sequences suggested that HWgs001 accounted for up to 93% of the gill microbiota. Microscopic observations suggested that HWgs001 lived within the gill tissue. Unlike symbionts of other bivalves, HWgs001 belongs to Alphaproteobacteria rather than Gammaproteobacteria, and no genes for carbon fixation were identified in its small genome. Moreover, HWgs001 was found to possess a dddP gene, responsible for the lysis of DMSP to acrylate. The enzymatic activity of dddP was confirmed using the heterologous expression, and in situ transcription of the gene in scallop gill tissues was demonstrated using reverse-transcription PCR. Together, these results revealed a taxonomically and functionally unique symbiont, which represents the first-documented DMSP-metabolizing symbiont likely to play significant roles in coastal marine ecosystems.
Keywords: Alphaproteobacteria; DMSP lyases; dddP; scallop; symbiont.
© 2023 The Authors. mLife published by John Wiley & Sons Australia, Ltd. on behalf of Institute of Microbiology, Chinese Academy of Sciences.
Conflict of interest statement
The authors declare no conflict of interests.
Figures



Similar articles
-
A novel ATP dependent dimethylsulfoniopropionate lyase in bacteria that releases dimethyl sulfide and acryloyl-CoA.Elife. 2021 May 10;10:e64045. doi: 10.7554/eLife.64045. Elife. 2021. PMID: 33970104 Free PMC article.
-
Oceanospirillales containing the DMSP lyase DddD are key utilisers of carbon from DMSP in coastal seawater.Microbiome. 2022 Jul 27;10(1):110. doi: 10.1186/s40168-022-01304-0. Microbiome. 2022. PMID: 35883169 Free PMC article.
-
The dddP gene of Roseovarius nubinhibens encodes a novel lyase that cleaves dimethylsulfoniopropionate into acrylate plus dimethyl sulfide.Microbiology (Reading). 2010 Jun;156(Pt 6):1900-1906. doi: 10.1099/mic.0.038927-0. Epub 2010 Apr 8. Microbiology (Reading). 2010. PMID: 20378650
-
Evolution of Dimethylsulfoniopropionate Metabolism in Marine Phytoplankton and Bacteria.Front Microbiol. 2017 Apr 19;8:637. doi: 10.3389/fmicb.2017.00637. eCollection 2017. Front Microbiol. 2017. PMID: 28469605 Free PMC article. Review.
-
Enzymology of Microbial Dimethylsulfoniopropionate Catabolism.Adv Protein Chem Struct Biol. 2017;109:195-222. doi: 10.1016/bs.apcsb.2017.05.001. Epub 2017 Jun 23. Adv Protein Chem Struct Biol. 2017. PMID: 28683918 Review.
Cited by
-
Scallop-bacteria symbiosis from the deep sea reveals strong genomic coupling in the absence of cellular integration.ISME J. 2024 Jan 8;18(1):wrae048. doi: 10.1093/ismejo/wrae048. ISME J. 2024. PMID: 38531780 Free PMC article.
-
Biofilm formation stabilizes metabolism in a Roseobacteraceae bacterium under temperature increase.Appl Environ Microbiol. 2023 Oct 31;89(10):e0060123. doi: 10.1128/aem.00601-23. Epub 2023 Sep 28. Appl Environ Microbiol. 2023. PMID: 37768087 Free PMC article.
References
-
- Kiene RP, Linn LJ, Bruton JA. New and important roles for DMSP in marine microbial communities. J Sea Res. 2000;43:209–24.
-
- Cirri E, Pohnert G. Algae‐bacteria interactions that balance the planktonic microbiome. New Phytol. 2019;223:100–6. - PubMed
-
- Zinke L. Cool gas in warm summers. Nat Clim Change. 2019;9:434.
-
- Dey M. Enzymology of microbial dimethylsulfoniopropionate catabolism. Adv Protein Chem Struct Biol. 2017;109:195–222. - PubMed
LinkOut - more resources
Full Text Sources
