Exploring the role of innate immunity in Cholangiocarcinoma: implications for prognosis, immune infiltration, and tumor metastasis
- PMID: 38817870
- PMCID: PMC11134439
- DOI: 10.7150/jca.94194
Exploring the role of innate immunity in Cholangiocarcinoma: implications for prognosis, immune infiltration, and tumor metastasis
Abstract
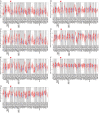
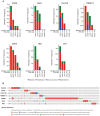
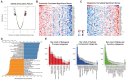
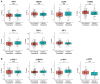
The innate immune system serves as the body's primary physiological defense against the intrusion of pathogenic microorganisms, playing a pivotal role in restricting viral infections. However, current research on the interplay between innate immune pathways and cancer is limited, with reported effects often inconsistent. Therefore, we aimed to elucidate the relationship between innate immune pathways and tumors through an amalgamation of bioinformatics and extensive data analysis. Conducting a pan-cancer analysis encompassing expression, genomic alterations, and clinical prognosis, we identified a close association between the innate immune pathway and cholangiocarcinoma. Subsequently, our focus shifted to unraveling the role of innate immune pathway proteins in cholangiocarcinoma. TIMER database analysis showed that the innate immune pathway predominantly influences the infiltration of macrophages and B cells in cholangiocarcinoma. Additionally, gene ontology (GO) and pathway analyses were performed for significantly differentially expressed genes correlated with the innate immune pathway in cholangiocarcinoma. Single-cell transcriptome analysis in cholangiocarcinoma demonstrated that genes in the innate immune pathway are primarily expressed in malignant cells, endothelial cells, monocytes and macrophages. To further validate the expression of proteins in the innate immune pathway in the tumor tissues of patients with cholangiocarcinoma, tumor tissue slices from patients with liver intrahepatic cholangiocarcinoma and normal tissue slices from the HPA database were analyzed. These results indicated pronounced activation of the innate immune pathway in the tumor tissues of patients with cholangiocarcinoma. Finally, proteomic data from patients with or without intrahepatic cholangiocarcinoma metastasis were analyzed. The results revealed a significant correlation between the expression and phosphorylation of IKKε and the occurrence of intrahepatic cholangiocarcinoma metastasis. These findings not only demonstrate the significance of the innate immune pathway in cholangiocarcinoma but also its potential as a prospective prognostic biomarker and therapeutic target for this malignancy.
Keywords: cholangiocarcinoma; immune cell infiltration; innate immune pathway; metastasis; prognosis.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures








References
-
- Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30:16–34. - PubMed
-
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. - PubMed
-
- Fischer S. Pattern Recognition Receptors and Control of Innate Immunity: Role of Nucleic Acids. Curr Pharm Biotechnol. 2018;19:1203–9. - PubMed
-
- Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol. 2014;32:461–88. - PubMed
LinkOut - more resources
Full Text Sources

