Virus versus host: influenza A virus circumvents the immune responses
- PMID: 38817972
- PMCID: PMC11137263
- DOI: 10.3389/fmicb.2024.1394510
Virus versus host: influenza A virus circumvents the immune responses
Abstract
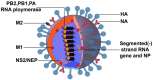
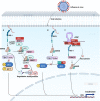
Influenza A virus (IAV) is a highly contagious pathogen causing dreadful losses to humans and animals around the globe. As is known, immune escape is a strategy that benefits the proliferation of IAVs by antagonizing, blocking, and suppressing immune surveillance. The HA protein binds to the sialic acid (SA) receptor to enter the cytoplasm and initiate viral infection. The conserved components of the viral genome produced during replication, known as the pathogen-associated molecular patterns (PAMPs), are thought to be critical factors for the activation of effective innate immunity by triggering dependent signaling pathways after recognition by pattern recognition receptors (PRRs), followed by a cascade of adaptive immunity. Viral infection-induced immune responses establish an antiviral state in the host to effectively inhibit virus replication and enhance viral clearance. However, IAV has evolved multiple mechanisms that allow it to synthesize and transport viral components by "playing games" with the host. At its heart, this review will describe how host and viral factors interact to facilitate the viral evasion of host immune responses.
Keywords: adaptive immunity; host-virus interaction; immune escape; influenza A virus; innate immunity.
Copyright © 2024 Su, Chen, Li and Shao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Induction of innate immunity and its perturbation by influenza viruses.Protein Cell. 2015 Oct;6(10):712-21. doi: 10.1007/s13238-015-0191-z. Epub 2015 Jul 24. Protein Cell. 2015. PMID: 26206138 Free PMC article. Review.
-
Protein Tyrosine Phosphatase SHP2 Suppresses Host Innate Immunity against Influenza A Virus by Regulating EGFR-Mediated Signaling.J Virol. 2021 Feb 24;95(6):e02001-20. doi: 10.1128/JVI.02001-20. Print 2021 Feb 24. J Virol. 2021. PMID: 33361428 Free PMC article.
-
Innate and adaptive immune responses against Influenza A Virus: Immune evasion and vaccination strategies.Immunobiology. 2022 Nov;227(6):152279. doi: 10.1016/j.imbio.2022.152279. Epub 2022 Sep 14. Immunobiology. 2022. PMID: 36272344 Review.
-
A host susceptibility gene, DR1, facilitates influenza A virus replication by suppressing host innate immunity and enhancing viral RNA replication.J Virol. 2015 Apr;89(7):3671-82. doi: 10.1128/JVI.03610-14. Epub 2015 Jan 14. J Virol. 2015. PMID: 25589657 Free PMC article.
-
Hemagglutinin of Influenza A Virus Antagonizes Type I Interferon (IFN) Responses by Inducing Degradation of Type I IFN Receptor 1.J Virol. 2015 Dec 16;90(5):2403-17. doi: 10.1128/JVI.02749-15. J Virol. 2015. PMID: 26676772 Free PMC article.
Cited by
-
Advancing RNA phage biology through meta-omics.Nucleic Acids Res. 2025 Apr 22;53(8):gkaf314. doi: 10.1093/nar/gkaf314. Nucleic Acids Res. 2025. PMID: 40263712 Free PMC article. Review.
-
Respiratory Virus-Specific and Time-Dependent Interference of Adenovirus Type 2, SARS-CoV-2 and Influenza Virus H1N1pdm09 During Viral Dual Co-Infection and Superinfection In Vitro.Viruses. 2024 Dec 19;16(12):1947. doi: 10.3390/v16121947. Viruses. 2024. PMID: 39772252 Free PMC article.
-
Immunological drivers of zoonotic virus emergence, evolution, and endemicity.Immunity. 2025 Apr 8;58(4):784-796. doi: 10.1016/j.immuni.2025.03.014. Epub 2025 Mar 31. Immunity. 2025. PMID: 40168990 Review.
-
Blood‑brain barrier dysfunction in schizophrenia: Mechanisms and implications (Review).Int J Mol Med. 2025 Oct;56(4):153. doi: 10.3892/ijmm.2025.5594. Epub 2025 Jul 25. Int J Mol Med. 2025. PMID: 40709398 Free PMC article. Review.
References
-
- Alymova I. V., Cipollo J. F., Parsons L. M., Music N., Kamal R. P., Tzeng W. P., et al. . (2022). Aberrant cellular glycosylation may increase the ability of influenza viruses to escape host immune responses through modification of the viral Glycome. MBio 13, e02983–e02921. doi: 10.1128/mbio.02983-21 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

