Application of whole exome sequencing in the diagnosis of muscular disorders: a study of Taiwanese pediatric patients
- PMID: 38818036
- PMCID: PMC11137626
- DOI: 10.3389/fgene.2024.1365729
Application of whole exome sequencing in the diagnosis of muscular disorders: a study of Taiwanese pediatric patients
Abstract
Background: Muscular dystrophies and congenital myopathies encompass various inherited muscular disorders that present diagnostic challenges due to clinical complexity and genetic heterogeneity.
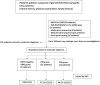
Methods: This study aimed to investigate the use of whole exome sequencing (WES) in diagnosing muscular disorders in pediatric patients in Taiwan. Out of 161 pediatric patients suspected to have genetic/inherited myopathies, 115 received a molecular diagnosis through conventional tests, single gene testing, and gene panels. The remaining 46 patients were divided into three groups: Group 1 (multiplex ligation-dependent probe amplification-negative Duchenne muscular dystrophy) with three patients (6.5%), Group 2 (various forms of muscular dystrophies) with 21 patients (45.7%), and Group 3 (congenital myopathies) with 22 patients (47.8%).
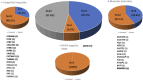
Results: WES analysis of these groups found pathogenic variants in 100.0% (3/3), 57.1% (12/21), and 68.2% (15/22) of patients in Groups 1 to 3, respectively. WES had a diagnostic yield of 65.2% (30 patients out of 46), detecting 30 pathogenic or potentially pathogenic variants across 28 genes.
Conclusion: WES enables the diagnosis of rare diseases with symptoms and characteristics similar to congenital myopathies and muscular dystrophies, such as muscle weakness. Consequently, this approach facilitates targeted therapy implementation and appropriate genetic counseling.
Keywords: congenital myopathies; muscular disorders; muscular dystrophies; mutational diversity; whole exome sequencing.
Copyright © 2024 Lee, Chuang, Chiu, Chang, Tu, Lo, Lin and Lin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Utilisation of exome sequencing for muscular disorders in Thai paediatric patients: diagnostic yield and mutational spectrum.Sci Rep. 2023 Jan 25;13(1):1376. doi: 10.1038/s41598-023-28405-6. Sci Rep. 2023. PMID: 36697461 Free PMC article.
-
Whole-Exome Sequencing Identifies Small Mutations in Pakistani Muscular Dystrophy Patients.Genet Test Mol Biomarkers. 2021 Mar;25(3):218-226. doi: 10.1089/gtmb.2020.0246. Genet Test Mol Biomarkers. 2021. PMID: 33734897
-
Use of Whole-Exome Sequencing for Diagnosis of Limb-Girdle Muscular Dystrophy: Outcomes and Lessons Learned.JAMA Neurol. 2015 Dec;72(12):1424-32. doi: 10.1001/jamaneurol.2015.2274. JAMA Neurol. 2015. PMID: 26436962
-
Prenatal diagnosis of congenital myopathies and muscular dystrophies.Clin Genet. 2016 Sep;90(3):199-210. doi: 10.1111/cge.12801. Epub 2016 Jun 2. Clin Genet. 2016. PMID: 27197572 Review.
-
Congenital myopathies and congenital muscular dystrophies.Curr Opin Neurol. 2001 Oct;14(5):575-82. doi: 10.1097/00019052-200110000-00005. Curr Opin Neurol. 2001. PMID: 11562568 Review.
Cited by
-
Compound Heterozygous RYR1 Variants in a Patient with Severe Congenital Myopathy: Case Report and Comparison with Additional Cases of Recessive RYR1-Related Myopathy.Int J Mol Sci. 2024 Oct 9;25(19):10867. doi: 10.3390/ijms251910867. Int J Mol Sci. 2024. PMID: 39409197 Free PMC article.
References
-
- Amendola L. M., Jarvik G. P., Leo M. C., McLaughlin H. M., Akkari Y., Amaral M. D., et al. (2016). Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the clinical sequencing exploratory research consortium. Am. J. Hum. Genet. 99, 247–276. 10.1016/j.ajhg.2016.06.001 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

