Proline Isomerization and Molten Globular Property of TgPDCD5 Secreted from Toxoplasma gondii Confers Its Regulation of Heparin Sulfate Binding
- PMID: 38818051
- PMCID: PMC11134355
- DOI: 10.1021/jacsau.3c00577
Proline Isomerization and Molten Globular Property of TgPDCD5 Secreted from Toxoplasma gondii Confers Its Regulation of Heparin Sulfate Binding
Abstract
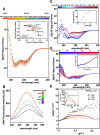
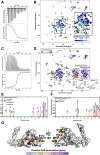
Toxoplasmosis, caused by Toxoplasma gondii, poses risks to vulnerable populations. TgPDCD5, a secreted protein of T. gondii, induces apoptosis through heparan sulfate-mediated endocytosis. The entry mechanism of TgPDCD5 has remained elusive. Here, we present the solution structure of TgPDCD5 as a helical bundle with an extended N-terminal helix, exhibiting molten globule characteristics. NMR perturbation studies reveal heparin/heparan sulfate binding involving the heparan sulfate/heparin proteoglycans-binding motif and the core region, influenced by proline isomerization of P107 residue. The heterogeneous proline recruits a cyclophilin TgCyp18, accelerating interconversion between conformers and regulating heparan/heparin binding. These atomic-level insights elucidate the binary switch's functionality, expose novel heparan sulfate-binding surfaces, and illuminate the unconventional cellular entry of pathogenic TgPDCD5.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




Similar articles
-
Overproduction of the pro-apoptotic molecule, programmed cell death 5, in Toxoplasma gondii leads to increased apoptosis of host macrophages.J Vet Med Sci. 2009 Sep;71(9):1183-9. doi: 10.1292/jvms.71.1183. J Vet Med Sci. 2009. PMID: 19801898
-
Programmed Cell Death 5 from Toxoplasma gondii: a secreted molecule that exerts a pro-apoptotic effect on host cells.Mol Biochem Parasitol. 2008 Jun;159(2):112-20. doi: 10.1016/j.molbiopara.2008.02.012. Epub 2008 Mar 6. Mol Biochem Parasitol. 2008. PMID: 18406478
-
NMR resonance assignments of the programmed cell death protein 5 (PDCD5) from Toxoplasma gondii.Biomol NMR Assign. 2020 Oct;14(2):277-280. doi: 10.1007/s12104-020-09961-8. Epub 2020 Jun 23. Biomol NMR Assign. 2020. PMID: 32578164
-
Conformation and dynamics of heparin and heparan sulfate.Glycobiology. 2000 Nov;10(11):1147-56. doi: 10.1093/glycob/10.11.1147. Glycobiology. 2000. PMID: 11087707 Review.
-
[Exploration of the molecular mechanism of ocular development and the creation of animal models for ocular diseases].Nippon Ganka Gakkai Zasshi. 2010 Mar;114(3):280-96; discussion 297. Nippon Ganka Gakkai Zasshi. 2010. PMID: 20387539 Review. Japanese.
References
-
- Morjaria S.; Epstein D. J.; Romero F. A.; Taur Y.; Seo S. K.; Papanicolaou G. A.; Hatzoglou V.; Rosenblum M.; Perales M. A.; Scordo M.; Kaltsas A. Toxoplasma Encephalitis in Atypical Hosts at an Academic Cancer Center.. Open Forum Infect Dis 2016, 3 (2), ofw070.10.1093/ofid/ofw070. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
