Changes in Active Site Loop Conformation Relate to the Transition toward a Novel Enzymatic Activity
- PMID: 38818068
- PMCID: PMC11134384
- DOI: 10.1021/jacsau.4c00179
Changes in Active Site Loop Conformation Relate to the Transition toward a Novel Enzymatic Activity
Abstract
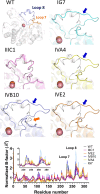
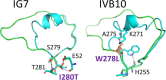
Enzymatic promiscuity, the ability of enzymes to catalyze multiple, distinct chemical reactions, has been well documented and is hypothesized to be a major driver of the emergence of new enzymatic functions. Yet, the molecular mechanisms involved in the transition from one activity to another remain debated and elusive. Here, we evaluated the redesign of the active site binding cleft of lactonase SsoPox using structure-based design and combinatorial libraries. We created variants with largely improved catalytic abilities against phosphotriesters, the best ones being >1000-fold better compared to the wild-type enzyme. The observed shifts in activity specificity are large, and some variants completely lost their initial activity. The selected combinations of mutations have considerably reshaped the active site cavity via side chain changes but mostly through large rearrangements of the active site loops and changes to their conformations, as revealed by a suite of crystal structures. This suggests that a specific active site loop configuration is critical to the lactonase activity. Interestingly, analysis of high-resolution structures hints at the potential role of conformational sampling and its directionality in defining the enzyme activity profile.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare the following competing financial interest(s): DD is a shareholder and CEO of Gene&GreenTK. EC is a shareholder and a co-founder of Gene&GreenTK. PJ, RB and EC report receiving personal fees from Gene&GreenTK during the study. MHE, DD and EC have filed the patent EP3941206. MHE and CB have a patent WO2020185861A1. MHE is a co-founder, a former CEO and an equity holder of Gene&GreenTK, a company that holds the license to WO2014167140 A1, FR 3068989 A1, FR 19/02834. MHE received fees from Gene&GreenTK. MHE interests with Gene&GreenTK have been reviewed and managed by the University of Minnesota in accordance with its Conflict-of-Interest policies. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Update of
-
Changes in Active Site Loop Conformation Relate to the Transition toward a Novel Enzymatic Activity.bioRxiv [Preprint]. 2023 May 26:2023.05.22.541809. doi: 10.1101/2023.05.22.541809. bioRxiv. 2023. Update in: JACS Au. 2024 Apr 25;4(5):1941-1953. doi: 10.1021/jacsau.4c00179. PMID: 37292757 Free PMC article. Updated. Preprint.
Similar articles
-
Changes in Active Site Loop Conformation Relate to the Transition toward a Novel Enzymatic Activity.bioRxiv [Preprint]. 2023 May 26:2023.05.22.541809. doi: 10.1101/2023.05.22.541809. bioRxiv. 2023. Update in: JACS Au. 2024 Apr 25;4(5):1941-1953. doi: 10.1021/jacsau.4c00179. PMID: 37292757 Free PMC article. Updated. Preprint.
-
Differential active site loop conformations mediate promiscuous activities in the lactonase SsoPox.PLoS One. 2013 Sep 23;8(9):e75272. doi: 10.1371/journal.pone.0075272. eCollection 2013. PLoS One. 2013. PMID: 24086491 Free PMC article.
-
Catalytic versatility and backups in enzyme active sites: the case of serum paraoxonase 1.J Mol Biol. 2012 May 4;418(3-4):181-96. doi: 10.1016/j.jmb.2012.02.042. Epub 2012 Mar 1. J Mol Biol. 2012. PMID: 22387469
-
Enzyme dynamics from NMR spectroscopy.Acc Chem Res. 2015 Feb 17;48(2):457-65. doi: 10.1021/ar500340a. Epub 2015 Jan 9. Acc Chem Res. 2015. PMID: 25574774 Free PMC article. Review.
-
Hybrid schemes based on quantum mechanics/molecular mechanics simulations goals to success, problems, and perspectives.Adv Protein Chem Struct Biol. 2011;85:81-142. doi: 10.1016/B978-0-12-386485-7.00003-X. Adv Protein Chem Struct Biol. 2011. PMID: 21920322 Review.
Cited by
-
Tuning the Envelope Structure of Enzyme Nanoreactors for In Vivo Detoxification of Organophosphates.Int J Mol Sci. 2023 Oct 30;24(21):15756. doi: 10.3390/ijms242115756. Int J Mol Sci. 2023. PMID: 37958742 Free PMC article.
-
Applications of Microbial Organophosphate-Degrading Enzymes to Detoxification of Organophosphorous Compounds for Medical Countermeasures against Poisoning and Environmental Remediation.Int J Mol Sci. 2024 Jul 17;25(14):7822. doi: 10.3390/ijms25147822. Int J Mol Sci. 2024. PMID: 39063063 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
