Deconvolution analysis identified altered hepatic cell landscape in primary sclerosing cholangitis and primary biliary cholangitis
- PMID: 38818402
- PMCID: PMC11138208
- DOI: 10.3389/fmed.2024.1327973
Deconvolution analysis identified altered hepatic cell landscape in primary sclerosing cholangitis and primary biliary cholangitis
Abstract
Introduction: Primary sclerosing cholangitis (PSC) and primary biliary cholangitis (PBC) are characterized by ductular reaction, hepatic inflammation, and liver fibrosis. Hepatic cells are heterogeneous, and functional roles of different hepatic cell phenotypes are still not defined in the pathophysiology of cholangiopathies. Cell deconvolution analysis estimates cell fractions of different cell phenotypes in bulk transcriptome data, and CIBERSORTx is a powerful deconvolution method to estimate cell composition in microarray data. CIBERSORTx performs estimation based on the reference file, which is referred to as signature matrix, and allows users to create custom signature matrix to identify specific phenotypes. In the current study, we created two custom signature matrices using two single cell RNA sequencing data of hepatic cells and performed deconvolution for bulk microarray data of liver tissues including PSC and PBC patients.
Methods: Custom signature matrix files were created using single-cell RNA sequencing data downloaded from GSE185477 and GSE115469. Custom signature matrices were validated for their deconvolution performance using validation data sets. Cell composition of each hepatic cell phenotype in the liver, which was identified in custom signature matrices, was calculated by CIBERSORTx and bulk RNA sequencing data of GSE159676. Deconvolution results were validated by analyzing marker expression for the cell phenotype in GSE159676 data.
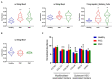
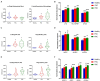
Results: CIBERSORTx and custom signature matrices showed comprehensive performance in estimation of population of various hepatic cell phenotypes. We identified increased population of large cholangiocytes in PSC and PBC livers, which is in agreement with previous studies referred to as ductular reaction, supporting the effectiveness and reliability of deconvolution analysis in this study. Interestingly, we identified decreased population of small cholangiocytes, periportal hepatocytes, and interzonal hepatocytes in PSC and PBC liver tissues compared to healthy livers.
Discussion: Although further studies are required to elucidate the roles of these hepatic cell phenotypes in cholestatic liver injury, our approach provides important implications that cell functions may differ depending on phenotypes, even in the same cell type during liver injury. Deconvolution analysis using CIBERSORTx could provide a novel approach for studies of specific hepatic cell phenotypes in liver diseases.
Keywords: CIBERSORTx; deconvolution analysis; hepatic cell landscape; primary biliary cholangitis; primary sclerosing cholangitis.
Copyright © 2024 Pham, Pham and Sato.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Liver transcriptome analysis reveals PSC-attributed gene set associated with fibrosis progression.JHEP Rep. 2024 Nov 12;7(3):101267. doi: 10.1016/j.jhepr.2024.101267. eCollection 2025 Mar. JHEP Rep. 2024. PMID: 39996122 Free PMC article.
-
Loss of apical sodium bile acid transporter alters bile acid circulation and reduces biliary damage in cholangitis.Am J Physiol Gastrointest Liver Physiol. 2023 Jan 1;324(1):G60-G77. doi: 10.1152/ajpgi.00112.2022. Epub 2022 Nov 21. Am J Physiol Gastrointest Liver Physiol. 2023. PMID: 36410025 Free PMC article.
-
Hepatic Stem/Progenitor Cell Activation Differs between Primary Sclerosing and Primary Biliary Cholangitis.Am J Pathol. 2018 Mar;188(3):627-639. doi: 10.1016/j.ajpath.2017.11.010. Epub 2017 Dec 15. Am J Pathol. 2018. PMID: 29248458
-
Macrophages in cholangiopathies.Curr Opin Gastroenterol. 2022 Mar 1;38(2):114-120. doi: 10.1097/MOG.0000000000000814. Curr Opin Gastroenterol. 2022. PMID: 35098932 Review.
-
Autoimmune biliary diseases: primary biliary cholangitis and primary sclerosing cholangitis.Pathologica. 2021 Jun;113(3):170-184. doi: 10.32074/1591-951X-245. Pathologica. 2021. PMID: 34294935 Free PMC article. Review.
Cited by
-
Cell type heterogeneity in gene co-expression networks: implications for toxicological research.Brief Bioinform. 2025 Jul 2;26(4):bbaf421. doi: 10.1093/bib/bbaf421. Brief Bioinform. 2025. PMID: 40856524 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

