Association of maternal genetics with the gut microbiome and eucalypt diet selection in captive koalas
- PMID: 38818452
- PMCID: PMC11138522
- DOI: 10.7717/peerj.17385
Association of maternal genetics with the gut microbiome and eucalypt diet selection in captive koalas
Abstract
Background: Koalas, an Australian arboreal marsupial, depend on eucalypt tree leaves for their diet. They selectively consume only a few of the hundreds of available eucalypt species. Since the koala gut microbiome is essential for the digestion and detoxification of eucalypts, their individual differences in the gut microbiome may lead to variations in their eucalypt selection and eucalypt metabolic capacity. However, research focusing on the relationship between the gut microbiome and differences in food preferences is very limited. We aimed to determine whether individual and regional differences exist in the gut microbiome of koalas as well as the mechanism by which these differences influence eucalypt selection.
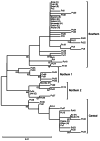
Methods: Foraging data were collected from six koalas and a total of 62 feces were collected from 15 koalas of two zoos in Japan. The mitochondrial phylogenetic analysis was conducted to estimate the mitochondrial maternal origin of each koala. In addition, the 16S-based gut microbiome of 15 koalas was analyzed to determine the composition and diversity of each koala's gut microbiome. We used these data to investigate the relationship among mitochondrial maternal origin, gut microbiome and eucalypt diet selection.
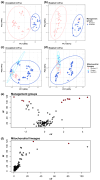
Results and discussion: This research revealed that diversity and composition of the gut microbiome and that eucalypt diet selection of koalas differs among regions. We also revealed that the gut microbiome alpha diversity was correlated with foraging diversity in koalas. These individual and regional differences would result from vertical (maternal) transmission of the gut microbiome and represent an intraspecific variation in koala foraging strategies. Further, we demonstrated that certain gut bacteria were strongly correlated with both mitochondrial maternal origin and eucalypt foraging patterns. Bacteria found to be associated with mitochondrial maternal origin included bacteria involved in fiber digestion and degradation of secondary metabolites, such as the families Rikenellaceae and Synergistaceae. These bacteria may cause differences in metabolic capacity between individual and regional koalas and influence their eucalypt selection.
Conclusion: We showed that the characteristics (composition and diversity) of the gut microbiome and eucalypt diet selection of koalas differ by individuals and regional origins as we expected. In addition, some gut bacteria that could influence eucalypt foraging of koalas showed the relationships with both mitochondrial maternal origin and eucalypt foraging pattern. These differences in the gut microbiome between regional origins may make a difference in eucalypt selection. Given the importance of the gut microbiome to koalas foraging on eucalypts and their strong symbiotic relationship, future studies should focus on the symbiotic relationship and coevolution between koalas and the gut microbiome to understand individual and regional differences in eucalypt diet selection by koalas.
Keywords: Captive animal; Diet selection; Diet specialist; Eucalyptus; Gut microbiome; Hindgut fermentor; Koala; Marsupial; Maternal transmission; Mitochondrial DNA.
©2024 Kondo et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures


Similar articles
-
The koala gut microbiome is largely unaffected by host translocation but rather influences host diet.Front Microbiol. 2023 Mar 2;14:1085090. doi: 10.3389/fmicb.2023.1085090. eCollection 2023. Front Microbiol. 2023. PMID: 36937253 Free PMC article.
-
Maternal inheritance of the koala gut microbiome and its compositional and functional maturation during juvenile development.Environ Microbiol. 2022 Jan;24(1):475-493. doi: 10.1111/1462-2920.15858. Epub 2021 Dec 9. Environ Microbiol. 2022. PMID: 34863030
-
Ingestion and Absorption of Eucalypt Monoterpenes in the Specialist Feeder, the Koala (Phascolarctos cinereus).J Chem Ecol. 2019 Sep;45(9):798-807. doi: 10.1007/s10886-019-01097-x. Epub 2019 Aug 17. J Chem Ecol. 2019. PMID: 31422515
-
Helping koalas battle disease - Recent advances in Chlamydia and koala retrovirus (KoRV) disease understanding and treatment in koalas.FEMS Microbiol Rev. 2020 Sep 1;44(5):583-605. doi: 10.1093/femsre/fuaa024. FEMS Microbiol Rev. 2020. PMID: 32556174 Free PMC article. Review.
-
Global landscape of gut microbiome diversity and antibiotic resistomes across vertebrates.Sci Total Environ. 2022 Sep 10;838(Pt 2):156178. doi: 10.1016/j.scitotenv.2022.156178. Epub 2022 May 23. Sci Total Environ. 2022. PMID: 35618126 Review.
Cited by
-
The Oral Microbiome in Queensland Free-Ranging Koalas (Phascolarctos cinereus) and Its Association with Age and Periodontal Disease.Animals (Basel). 2025 Jun 20;15(13):1834. doi: 10.3390/ani15131834. Animals (Basel). 2025. PMID: 40646733 Free PMC article.
References
-
- Allison MJ, Mayberry WR, Mcsweeney CS, Stahl DA. Synergistes jonesii, gen. nov., sp. nov.: a rumen bacterium that degrades toxic pyridinediols. Systematic and Applied Microbiology. 1992;15(4):522–529. doi: 10.1016/S0723-2020(11)80111-6. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources

