The chloroplast genome inheritance pattern of the Deli-Nigerian prospection material (NPM) × Yangambi population of Elaeis guineensis Jacq
- PMID: 38818457
- PMCID: PMC11138521
- DOI: 10.7717/peerj.17335
The chloroplast genome inheritance pattern of the Deli-Nigerian prospection material (NPM) × Yangambi population of Elaeis guineensis Jacq
Abstract
Background: The chloroplast genome has the potential to be genetically engineered to enhance the agronomic value of major crops. As a crop plant with major economic value, it is important to understand every aspect of the genetic inheritance pattern among Elaeis guineensis individuals to ensure the traceability of agronomic traits.
Methods: Two parental E. guineensis individuals and 23 of their F1 progenies were collected and sequenced using the next-generation sequencing (NGS) technique on the Illumina platform. Chloroplast genomes were assembled de novo from the cleaned raw reads and aligned to check for variations. The sequences were compared and analyzed with programming language scripting and relevant bioinformatic softwares. Simple sequence repeat (SSR) loci were determined from the chloroplast genome.
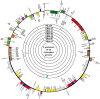
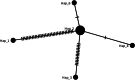
Results: The chloroplast genome assembly resulted in 156,983 bp, 156,988 bp, 156,982 bp, and 156,984 bp. The gene content and arrangements were consistent with the reference genome published in the GenBank database. Seventy-eight SSRs were detected in the chloroplast genome, with most located in the intergenic spacer region.The chloroplast genomes of 17 F1 progenies were exact copies of the maternal parent, while six individuals showed a single variation in the sequence. Despite the significant variation displayed by the male parent, all the nucleotide variations were synonymous. This study show highly conserve gene content and sequence in Elaeis guineensis chloroplast genomes. Maternal inheritance of chloroplast genome among F1 progenies are robust with a low possibility of mutations over generations. The findings in this study can enlighten inheritance pattern of Elaeis guineensis chloroplast genome especially among crops' scientists who consider using chloroplast genome for agronomic trait modifications.
Keywords: Chloroplast genome; Chloroplast inheritance; Gene content; Oil palm; Simple sequence repeat.
©2024 Mohd Talkah et al.
Conflict of interest statement
Muhammad Farid Abdul Rahim, Nurul Fatiha Farhana Hanafi & Mohd Azinuddin Ahmad Mokhtar are employees of FGV R&D Sdn Bhd/FGV Holdings, Malaysia. The authors declare there are no competing interests.
Figures

Similar articles
-
Characterization of the chloroplast genome sequence of oil palm (Elaeis guineensis Jacq.).Gene. 2012 Jun 1;500(2):172-80. doi: 10.1016/j.gene.2012.03.061. Epub 2012 Apr 2. Gene. 2012. PMID: 22487870
-
Genotyping by sequencing for the construction of oil palm (Elaeis guineensis Jacq.) genetic linkage map and mapping of yield related quantitative trait loci.PeerJ. 2024 Jan 30;12:e16570. doi: 10.7717/peerj.16570. eCollection 2024. PeerJ. 2024. PMID: 38313025 Free PMC article.
-
Exploiting transcriptome data for the development and characterization of gene-based SSR markers related to cold tolerance in oil palm (Elaeis guineensis).BMC Plant Biol. 2014 Dec 19;14:384. doi: 10.1186/s12870-014-0384-2. BMC Plant Biol. 2014. PMID: 25522814 Free PMC article.
-
Genetic architecture of palm oil fatty acid composition in cultivated oil palm (Elaeis guineensis Jacq.) compared to its wild relative E. oleifera (H.B.K) Cortés.PLoS One. 2014 May 9;9(5):e95412. doi: 10.1371/journal.pone.0095412. eCollection 2014. PLoS One. 2014. PMID: 24816555 Free PMC article.
-
Development and validation of whole genome-wide and genic microsatellite markers in oil palm (Elaeis guineensis Jacq.): First microsatellite database (OpSatdb).Sci Rep. 2019 Feb 13;9(1):1899. doi: 10.1038/s41598-018-37737-7. Sci Rep. 2019. PMID: 30760842 Free PMC article.
Cited by
-
The chloroplast genome of the allotriploid, Paeonia × lemoinei cv. Oukan, and its phylogenetic implications.Mitochondrial DNA B Resour. 2025 Jun 23;10(7):615-619. doi: 10.1080/23802359.2025.2519229. eCollection 2025. Mitochondrial DNA B Resour. 2025. PMID: 40589895 Free PMC article.
References
-
- Abdullah Shahzadi I, Mehmood F, Ali Z, Malik MS, Waseem S, Mirza B, Ahmed I, Waheed MT. Comparative analyses of chloroplast genomes among three Firmiana species: identification of mutational hotspots and phylogenetic relationship with other species of Malvaceae. Plant Gene. 2019;19:1–10. doi: 10.1016/j.plgene.2019.100199. - DOI
-
- Abubakar A, Ishak MY. An overview of the role of smallholders in oil palm production systems in changing climate. Nature Environment & Pollution Technology. 2022;21(5):2055–2071. doi: 10.46488/NEPT.2022.v21i05.004. - DOI
-
- Bansal KC, Saha D. Chloroplast genomics and genetic engineering for crop improvement. Agricultural Research. 2012;1:53–66. doi: 10.1007/s40003-011-0010-6. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

