Neutralizing gut-derived lipopolysaccharide as a novel therapeutic strategy for severe leptospirosis
- PMID: 38818711
- PMCID: PMC11142641
- DOI: 10.7554/eLife.96065
Neutralizing gut-derived lipopolysaccharide as a novel therapeutic strategy for severe leptospirosis
Abstract
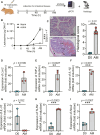
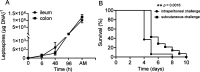
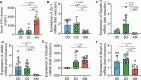
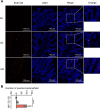
Leptospirosis is an emerging infectious disease caused by pathogenic Leptospira spp. Humans and some mammals can develop severe forms of leptospirosis accompanied by a dysregulated inflammatory response, which often results in death. The gut microbiota has been increasingly recognized as a vital element in systemic health. However, the precise role of the gut microbiota in severe leptospirosis is still unknown. Here, we aimed to explore the function and potential mechanisms of the gut microbiota in a hamster model of severe leptospirosis. Our study showed that leptospires were able to multiply in the intestine, cause pathological injury, and induce intestinal and systemic inflammatory responses. 16S rRNA gene sequencing analysis revealed that Leptospira infection changed the composition of the gut microbiota of hamsters with an expansion of Proteobacteria. In addition, gut barrier permeability was increased after infection, as reflected by a decrease in the expression of tight junctions. Translocated Proteobacteria were found in the intestinal epithelium of moribund hamsters, as determined by fluorescence in situ hybridization, with elevated lipopolysaccharide (LPS) levels in the serum. Moreover, gut microbiota depletion reduced the survival time, increased the leptospiral load, and promoted the expression of proinflammatory cytokines after Leptospira infection. Intriguingly, fecal filtration and serum from moribund hamsters both increased the transcription of TNF-α, IL-1β, IL-10, and TLR4 in macrophages compared with those from uninfected hamsters. These stimulating activities were inhibited by LPS neutralization using polymyxin B. Based on our findings, we identified an LPS neutralization therapy that significantly improved the survival rates in severe leptospirosis when used in combination with antibiotic therapy or polyclonal antibody therapy. In conclusion, our study not only uncovers the role of the gut microbiota in severe leptospirosis but also provides a therapeutic strategy for severe leptospirosis.
Keywords: Leptospira; hamster; infectious disease; microbiology; microbiota.
© 2024, Xie et al.
Conflict of interest statement
XX, XC, SZ, JL, WZ, YC No competing interests declared
Figures







Update of
- doi: 10.1101/2024.01.17.576119
- doi: 10.7554/eLife.96065.1
- doi: 10.7554/eLife.96065.2
Similar articles
-
Gut microbiota-derived butyrate improved acute leptospirosis in hamster via promoting macrophage ROS mediated by HDAC3 inhibition.mBio. 2024 Oct 16;15(10):e0190624. doi: 10.1128/mbio.01906-24. Epub 2024 Sep 17. mBio. 2024. PMID: 39287437 Free PMC article.
-
The pre-activated immune response induced by LPS protects host from leptospirosis.PLoS One. 2020 Nov 24;15(11):e0242742. doi: 10.1371/journal.pone.0242742. eCollection 2020. PLoS One. 2020. PMID: 33232366 Free PMC article.
-
Gut microbiota involved in leptospiral infections.ISME J. 2022 Mar;16(3):764-773. doi: 10.1038/s41396-021-01122-6. Epub 2021 Sep 29. ISME J. 2022. PMID: 34588617 Free PMC article.
-
Expression of TNF-alpha, TGF-beta, IP-10 and IL-10 mRNA in kidneys of hamsters infected with pathogenic Leptospira.Comp Immunol Microbiol Infect Dis. 2010 Sep;33(5):423-34. doi: 10.1016/j.cimid.2009.05.001. Epub 2009 Jun 25. Comp Immunol Microbiol Infect Dis. 2010. PMID: 19559480
-
Immune responses to Leptospira infection: roles as biomarkers for disease severity.Braz J Infect Dis. 2014 Jan-Feb;18(1):77-81. doi: 10.1016/j.bjid.2013.08.002. Epub 2013 Nov 22. Braz J Infect Dis. 2014. PMID: 24275371 Free PMC article. Review.
Cited by
-
Myocarditis and neutrophil-mediated vascular leakage but not cytokine storm associated with fatal murine leptospirosis.EBioMedicine. 2025 Feb;112:105571. doi: 10.1016/j.ebiom.2025.105571. Epub 2025 Jan 30. EBioMedicine. 2025. PMID: 39889371 Free PMC article.
References
-
- Alventosa Mateu C. Gastrointestinal bleeding and acute hepatic failure by leptospirosis: an entity that should not be forgotten. J Rev Gastroenterol Peru. 2017;37:96–99. - PubMed
-
- Becattini S, Sorbara MT, Kim SG, Littmann EL, Dong Q, Walsh G, Wright R, Amoretti L, Fontana E, Hohl TM, Pamer EG. Rapid transcriptional and metabolic adaptation of intestinal microbes to host immune activation. Cell Host & Microbe. 2021;29:378–393. doi: 10.1016/j.chom.2021.01.003. - DOI - PMC - PubMed
-
- Bernard-Raichon L, Venzon M, Klein J, Axelrad JE, Zhang C, Sullivan AP, Hussey GA, Casanovas-Massana A, Noval MG, Valero-Jimenez AM, Gago J, Putzel G, Pironti A, Wilder E, Yale IMPACT Research Team. Thorpe LE, Littman DR, Dittmann M, Stapleford KA, Shopsin B, Torres VJ, Ko AI, Iwasaki A, Cadwell K, Schluter J. Gut microbiome dysbiosis in antibiotic-treated COVID-19 patients is associated with microbial translocation and bacteremia. Nature Communications. 2022;13:5926. doi: 10.1038/s41467-022-33395-6. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

