Atg44/Mdi1/mitofissin facilitates Dnm1-mediated mitochondrial fission
- PMID: 38818923
- PMCID: PMC11423663
- DOI: 10.1080/15548627.2024.2360345
Atg44/Mdi1/mitofissin facilitates Dnm1-mediated mitochondrial fission
Abstract
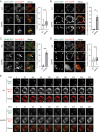
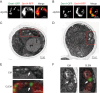
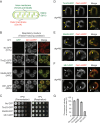
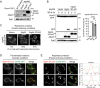
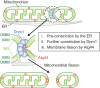
Mitochondria undergo fission and fusion, and their coordinated balance is crucial for maintaining mitochondrial homeostasis. In yeast, the dynamin-related protein Dnm1 is a mitochondrial fission factor acting from outside the mitochondria. We recently reported the mitochondrial intermembrane space protein Atg44/mitofissin/Mdi1/Mco8 as a novel fission factor, but the relationship between Atg44 and Dnm1 remains elusive. Here, we show that Atg44 is required to complete Dnm1-mediated mitochondrial fission under homeostatic conditions. Atg44-deficient cells often exhibit enlarged mitochondria with accumulated Dnm1 and rosary-like mitochondria with Dnm1 foci at constriction sites. These mitochondrial constriction sites retain the continuity of both the outer and inner membranes within an extremely confined space, indicating that Dnm1 is unable to complete mitochondrial fission without Atg44. Moreover, accumulated Atg44 proteins are observed at mitochondrial constriction sites. These findings suggest that Atg44 and Dnm1 cooperatively execute mitochondrial fission from inside and outside the mitochondria, respectively.Abbreviation: ATG: autophagy related; CLEM: correlative light and electron microscopy; EM: electron microscopy; ER: endoplasmic reticulum; ERMES: endoplasmic reticulum-mitochondria encounter structure; GA: glutaraldehyde; GFP: green fluorescent protein; GTP: guanosine triphosphate: IMM: inner mitochondrial membrane; IMS: intermembrane space; OMM: outer mitochondrial membrane; PB: phosphate buffer; PBS: phosphate-buffered saline; PFA: paraformaldehyde; RFP: red fluorescent protein; WT: wild type.
Keywords: Atg44; Dnm1; mitochondrial fission; mitofissin; mitophagy; yeast.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
