Comprehensive analysis of abnormal methylation modification differential expression mRNAs between low-grade and high-grade intervertebral disc degeneration and its correlation with immune cells
- PMID: 38819022
- PMCID: PMC11146251
- DOI: 10.1080/07853890.2024.2357742
Comprehensive analysis of abnormal methylation modification differential expression mRNAs between low-grade and high-grade intervertebral disc degeneration and its correlation with immune cells
Abstract
Background: Intervertebral disc degeneration (IDD) is an important cause of low back pain. The aim of this study is to identify the potential molecular mechanism of abnormal methylation-modified DNA in the progression of IDD, hoping to contribute to the diagnosis and management of IDD.
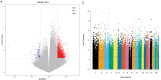
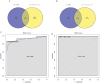
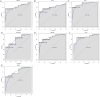
Methods: Low-grade IDD (grade I-II) and high-grade IDD (grade III-V) data were downloaded from GSE70362 and GSE129789 datasets. The abnormally methylated modified differentially expressed mRNAs (DEmRNAs) were identified by differential expression analysis (screening criteria were p < .05 and |logFC| > 1) and differential methylation analysis (screening criteria were p < .05 and |δβ| > 0.1). The classification models were constructed, and the receiver operating characteristic analysis was also carried out. In addition, functional enrichment analysis and immune correlation analysis were performed and the miRNAs targeted for the abnormally methylated DEmRNAs were predicted. Finally, expression validation was performed using real-time PCR.
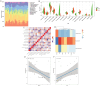
Results: Compared with low-grade IDD, seven abnormal methylation-modified DEmRNAs (AOX1, IBSP, QDPR, ABLIM1, CRISPLD2, ACTC1 and EMILIN1) were identified in high-grade IDD, and the classification models of random forests (RF) and support vector machine (SVM) were constructed. Moreover, seven abnormal methylation-modified DEmRNAs and classification models have high diagnostic accuracy (area under the curve [AUC] > 0.8). We also found that AUC values of single abnormal methylation-modified DEmRNA were all lower than those of RF and SVM classification models. Pearson correlation analysis found that macrophages M2 and EMILIN1 had significant negative correlation, while macrophages M2 and IBSP had significant positive correlation. In addition, four targeted relationship pairs (hsa-miR-4728-5p-QDPR, hsa-miR-4533-ABLIM1, hsa-miR-4728-5p-ABLIM1 and hsa-miR-4534-CRISPLD2) and multiple signalling pathways (for example, PI3K-AKT signalling pathway, osteoclast differentiation and calcium signalling pathway) were also identified that may be involved in the progression of IDD.
Conclusion: The identification of abnormal methylation-modified DEmRNAs and the construction of classification models in this study were helpful for the diagnosis and management of IDD progression.
Keywords: Intervertebral disc degeneration; biomarkers; diagnostic; mRNAs; methylation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Shi Q, Xu L, Zeng X.. Sirtuin 1 participates in intervertebral disc degeneration via the nicotinamide phosphoribosyl transferase/nicotinamide adenine dinucleotide/sirtuin 1 pathway responsible for regulating autophagy of nucleus pulposus cells. Exp Ther Med. 2022;23(4):267. doi: 10.3892/etm.2022.11193. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
