CRISPR Dependency Screens in Primary Hematopoietic Stem Cells Identify KDM3B as a Genotype-specific Vulnerability in IDH2- and TET2-mutant Cells
- PMID: 38819218
- PMCID: PMC11452290
- DOI: 10.1158/2159-8290.CD-23-1092
CRISPR Dependency Screens in Primary Hematopoietic Stem Cells Identify KDM3B as a Genotype-specific Vulnerability in IDH2- and TET2-mutant Cells
Abstract
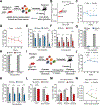
Clonal hematopoiesis (CH) is a common premalignant state in the blood and confers an increased risk of blood cancers and all-cause mortality. Identification of therapeutic targets in CH has been hindered by the lack of an ex vivo platform amenable for studying primary hematopoietic stem and progenitor cells (HSPCs). Here, we utilize an ex vivo co-culture system of HSPCs with bone marrow endothelial cells to perform CRISPR/Cas9 screens in mutant HSPCs. Our data reveal that loss of the histone demethylase family members Kdm3b and Jmjd1c specifically reduces the fitness of Idh2- and Tet2-mutant HSPCs. Kdm3b loss in mutant cells leads to decreased expression of critical cytokine receptors including Mpl, rendering mutant HSPCs preferentially susceptible to inhibition of downstream JAK2 signaling. Our study nominates an epigenetic regulator and an epigenetically regulated receptor signaling pathway as genotype-specific therapeutic targets and provides a scalable platform to identify genetic dependencies in mutant HSPCs. Significance: Given the broad prevalence, comorbidities, and risk of malignant transformation associated with CH, there is an unmet need to identify therapeutic targets. We develop an ex vivo platform to perform CRISPR/Cas9 screens in primary HSPCs. We identify KDM3B and downstream signaling components as genotype-specific dependencies in CH and myeloid malignancies. See related commentary by Khabusheva and Goodell, p. 1768.
©2024 American Association for Cancer Research.
Conflict of interest statement
Figures


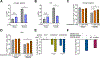

References
MeSH terms
Substances
Grants and funding
- R00 CA252005/CA/NCI NIH HHS/United States
- K08 CA241371/CA/NCI NIH HHS/United States
- R01 CA259273/CA/NCI NIH HHS/United States
- P01 CA108671/CA/NCI NIH HHS/United States
- P01 CA066996/CA/NCI NIH HHS/United States
- R01 CA176745/CA/NCI NIH HHS/United States
- Leukemia Research Foundation (LRF)
- 'la Caixa' Foundation ('la Caixa')
- Fonds de Recherche du Québec - Santé (FRQS)
- K99 CA248460/CA/NCI NIH HHS/United States
- Geoffrey Beene Foundation (GBF)
- P50 CA254838/CA/NCI NIH HHS/United States
- R01 CA260711/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- U01 AG077925/AG/NIA NIH HHS/United States
- F31 CA257367/CA/NCI NIH HHS/United States
- K99 CA252005/CA/NCI NIH HHS/United States
- F99 CA274656/CA/NCI NIH HHS/United States
- R35 CA197594/CA/NCI NIH HHS/United States
- K08 CA267058/CA/NCI NIH HHS/United States
- R00 CA248460/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

