SMAD4 promotes somatic-germline contact during murine oocyte growth
- PMID: 38819913
- PMCID: PMC11142639
- DOI: 10.7554/eLife.91798
SMAD4 promotes somatic-germline contact during murine oocyte growth
Abstract
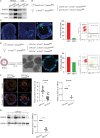
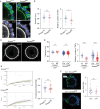
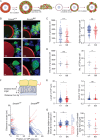
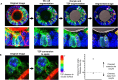
Development of the mammalian oocyte requires physical contact with the surrounding granulosa cells of the follicle, which provide it with essential nutrients and regulatory signals. This contact is achieved through specialized filopodia, termed transzonal projections (TZPs), that extend from the granulosa cells to the oocyte surface. Transforming growth factor (TGFβ) family ligands produced by the oocyte increase the number of TZPs, but how they do so is unknown. Using an inducible Cre recombinase strategy together with expression of green fluorescent protein to verify Cre activity in individual cells, we examined the effect of depleting the canonical TGFβ mediator, SMAD4, in mouse granulosa cells. We observed a 20-50% decrease in the total number of TZPs in SMAD4-depleted granulosa cell-oocyte complexes, and a 50% decrease in the number of newly generated TZPs when the granulosa cells were reaggregated with wild-type oocytes. Three-dimensional image analysis revealed that TZPs of SMAD4-depleted cells were longer than controls and more frequently oriented towards the oocyte. Strikingly, the transmembrane proteins, N-cadherin and Notch2, were reduced by 50% in SMAD4-depleted cells. SMAD4 may thus modulate a network of cell adhesion proteins that stabilize the attachment of TZPs to the oocyte, thereby amplifying signalling between the two cell types.
Keywords: SMAD; developmental biology; follicle; granulosa; mouse; oocyte; transzonal projections.
© 2024, Granados-Aparici et al.
Conflict of interest statement
SG, QY, HC No competing interests declared
Figures


Update of
- doi: 10.1101/2023.08.31.555753
- doi: 10.7554/eLife.91798.1
- doi: 10.7554/eLife.91798.2
Similar articles
-
MYO10 promotes transzonal projection-dependent germ line-somatic contact during mammalian folliculogenesis†.Biol Reprod. 2022 Aug 9;107(2):474-487. doi: 10.1093/biolre/ioac078. Biol Reprod. 2022. PMID: 35470858 Free PMC article.
-
Transzonal projections: Essential structures mediating intercellular communication in the mammalian ovarian follicle.Mol Reprod Dev. 2022 Nov;89(11):509-525. doi: 10.1002/mrd.23645. Epub 2022 Sep 16. Mol Reprod Dev. 2022. PMID: 36112806 Review.
-
Minimal fertility defects in mice deficient in oocyte-expressed Smad4.Biol Reprod. 2012 Jan 30;86(1):1-6. doi: 10.1095/biolreprod.111.094375. Print 2012 Jan. Biol Reprod. 2012. PMID: 21900682
-
Reestablishment of transzonal projections and growth of bovine oocytes in vitro.J Reprod Dev. 2021 Oct 29;67(5):300-306. doi: 10.1262/jrd.2021-036. Epub 2021 Aug 20. J Reprod Dev. 2021. PMID: 34421085 Free PMC article.
-
Regulation of germ cell development by intercellular signaling in the mammalian ovarian follicle.Wiley Interdiscip Rev Dev Biol. 2018 Jan;7(1):10.1002/wdev.294. doi: 10.1002/wdev.294. Epub 2017 Sep 11. Wiley Interdiscip Rev Dev Biol. 2018. PMID: 28892263 Free PMC article. Review.
Cited by
-
Reproductive health.Elife. 2025 Jan 29;14:e102432. doi: 10.7554/eLife.102432. Elife. 2025. PMID: 39878459 Free PMC article.
-
The role of lncRNA HUPCOS in androgen metabolism and follicle growth arrest in polycystic ovary syndrome.Cell Biol Toxicol. 2025 Jun 17;41(1):105. doi: 10.1007/s10565-025-10047-1. Cell Biol Toxicol. 2025. PMID: 40524087 Free PMC article.
-
Mechano-regulation of germline development, maintenance, and differentiation.BBA Adv. 2024 Nov 30;6:100127. doi: 10.1016/j.bbadva.2024.100127. eCollection 2024. BBA Adv. 2024. PMID: 39720163 Free PMC article.
References
-
- Aragón E, Wang Q, Zou Y, Morgani SM, Ruiz L, Kaczmarska Z, Su J, Torner C, Tian L, Hu J, Shu W, Agrawal S, Gomes T, Márquez JA, Hadjantonakis A-K, Macias MJ, Massagué J. Structural basis for distinct roles of SMAD2 and SMAD3 in FOXH1 pioneer-directed TGF-β signaling. Genes & Development. 2019;33:1506–1524. doi: 10.1101/gad.330837.119. - DOI - PMC - PubMed
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

