Information content and optimization of self-organized developmental systems
- PMID: 38819997
- PMCID: PMC11161761
- DOI: 10.1073/pnas.2322326121
Information content and optimization of self-organized developmental systems
Abstract
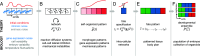
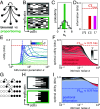
A key feature of many developmental systems is their ability to self-organize spatial patterns of functionally distinct cell fates. To ensure proper biological function, such patterns must be established reproducibly, by controlling and even harnessing intrinsic and extrinsic fluctuations. While the relevant molecular processes are increasingly well understood, we lack a principled framework to quantify the performance of such stochastic self-organizing systems. To that end, we introduce an information-theoretic measure for self-organized fate specification during embryonic development. We show that the proposed measure assesses the total information content of fate patterns and decomposes it into interpretable contributions corresponding to the positional and correlational information. By optimizing the proposed measure, our framework provides a normative theory for developmental circuits, which we demonstrate on lateral inhibition, cell type proportioning, and reaction-diffusion models of self-organization. This paves a way toward a classification of developmental systems based on a common information-theoretic language, thereby organizing the zoo of implicated chemical and mechanical signaling processes.
Keywords: development; information theory; self-organization; signaling networks.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures





Similar articles
-
Pattern formation mechanisms of self-organizing reaction-diffusion systems.Dev Biol. 2020 Apr 1;460(1):2-11. doi: 10.1016/j.ydbio.2019.10.031. Epub 2020 Jan 30. Dev Biol. 2020. PMID: 32008805 Free PMC article. Review.
-
Positional information, positional error, and readout precision in morphogenesis: a mathematical framework.Genetics. 2015 Jan;199(1):39-59. doi: 10.1534/genetics.114.171850. Epub 2014 Oct 31. Genetics. 2015. PMID: 25361898 Free PMC article.
-
AI-powered simulation-based inference of a genuinely spatial-stochastic gene regulation model of early mouse embryogenesis.PLoS Comput Biol. 2024 Nov 14;20(11):e1012473. doi: 10.1371/journal.pcbi.1012473. eCollection 2024 Nov. PLoS Comput Biol. 2024. PMID: 39541410 Free PMC article.
-
The dynamic transmission of positional information in stau- mutants during Drosophila embryogenesis.Elife. 2020 Jun 8;9:e54276. doi: 10.7554/eLife.54276. Elife. 2020. PMID: 32511091 Free PMC article.
-
Self-organized cell migration across scales - from single cell movement to tissue formation.Development. 2021 Apr 6;148(7):dev191767. doi: 10.1242/dev.191767. Development. 2021. PMID: 33824176 Review.
References
-
- Wolpert L., Positional information and the spatial pattern of cellular differentiation. J. Theor. Biol. 25, 1–47 (1969). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

