Bacterial vampirism mediated through taxis to serum
- PMID: 38820052
- PMCID: PMC11142651
- DOI: 10.7554/eLife.93178
Bacterial vampirism mediated through taxis to serum
Abstract
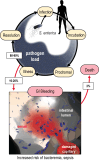
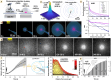
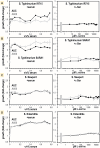
Bacteria of the family Enterobacteriaceae are associated with gastrointestinal (GI) bleeding and bacteremia and are a leading cause of death, from sepsis, for individuals with inflammatory bowel diseases. The bacterial behaviors and mechanisms underlying why these bacteria are prone to bloodstream entry remain poorly understood. Herein, we report that clinical isolates of non-typhoidal Salmonella enterica serovars, Escherichia coli, and Citrobacter koseri are rapidly attracted toward sources of human serum. To simulate GI bleeding, we utilized an injection-based microfluidics device and found that femtoliter volumes of human serum are sufficient to induce bacterial attraction to the serum source. This response is orchestrated through chemotaxis and the chemoattractant L-serine, an amino acid abundant in serum that is recognized through direct binding by the chemoreceptor Tsr. We report the first crystal structures of Salmonella Typhimurium Tsr in complex with L-serine and identify a conserved amino acid recognition motif for L-serine shared among Tsr orthologues. We find Tsr to be widely conserved among Enterobacteriaceae and numerous World Health Organization priority pathogens associated with bloodstream infections. Lastly, we find that Enterobacteriaceae use human serum as a source of nutrients for growth and that chemotaxis and the chemoreceptor Tsr provide a competitive advantage for migration into enterohemorrhagic lesions. We define this bacterial behavior of taxis toward serum, colonization of hemorrhagic lesions, and the consumption of serum nutrients as 'bacterial vampirism', which may relate to the proclivity of Enterobacteriaceae for bloodstream infections.
Keywords: E. coli; Salmonella; Tsr; chemoreceptor; chemotaxis; infectious disease; microbiology; molecular biophysics; salmonella enterica typhimurium; serine; serum; structural biology.
Plain language summary
Sepsis is the leading cause of death in patients with inflammatory bowel disease. Individuals with this condition can experience recurrent episodes of intestinal bleeding, giving intestinal (or enteric) bacteria an entry point into the bloodstream. This puts patients at risk of developing fatal infections – particularly from infections caused by bacteria belonging to the Enterobacteriaceae family. However, it is not well understood why this family of bacteria are particularly prone to entering the bloodstream. Enteric bacteria commonly respond to chemicals (or chemical stimuli) in their environment. This process, known as chemotaxis, helps bacteria with a variety of tasks, such as monitoring their environment, moving to different areas within their environment or colonizing their host. Chemical stimuli are classed as ‘attractants’ or ‘repellents’, with attractants luring the bacteria to an area and repellents discouraging the bacteria from being in a specific place. Intestinal bleeds will release serum (the liquid part of blood) into the gut, which could serve as a source of chemical stimuli to attract Enterobacteriaceae into the bloodstream. To find out more, Glenn, Gentry-Lear et al. first used a microfluidic device to simulate an intestinal bleed and tested the response of Enterobacteriaceae bacteria to serum. Using chemotaxis, bacteria were found to be attracted to the amino acid L-serine in the serum to which they were able to attach through a receptor called Tsr. They also consumed nutrients present in the human serum to help them grow. Experiments with intestinal tissue showed that chemotaxis attracted bacteria to bleeding blood vessels and the Tsr receptor helped them to infiltrate the blood vessels. Glenn et al. termed this attraction to and feeding upon blood serum as ‘bacterial vampirism’. These findings suggest that chemotaxis of Enterobacteriaceae towards L-serine in serum may be linked to their tendency to enter the bloodstream. Developing therapies that target chemotaxis in Enterobacteriaceae may provide a method for managing bloodstream infections.
© 2023, Glenn, Gentry-Lear et al.
Conflict of interest statement
SG, ZG, MS, MH, TA No competing interests declared, AB owns Amethyst Antimicrobials, LLC
Figures












Update of
-
Bacterial vampirism mediated through taxis to serum.bioRxiv [Preprint]. 2024 Jan 24:2023.07.07.548164. doi: 10.1101/2023.07.07.548164. bioRxiv. 2024. Update in: Elife. 2024 May 31;12:RP93178. doi: 10.7554/eLife.93178. PMID: 37461633 Free PMC article. Updated. Preprint.
Similar articles
-
Navigating contradictions in enteric chemotactic stimuli.Elife. 2025 Aug 11;14:RP106261. doi: 10.7554/eLife.106261. Elife. 2025. PMID: 40788306 Free PMC article.
-
Bacterial vampirism mediated through taxis to serum.bioRxiv [Preprint]. 2024 Jan 24:2023.07.07.548164. doi: 10.1101/2023.07.07.548164. bioRxiv. 2024. Update in: Elife. 2024 May 31;12:RP93178. doi: 10.7554/eLife.93178. PMID: 37461633 Free PMC article. Updated. Preprint.
-
Salmonella exploits host- and bacterial-derived β-alanine for replication inside host macrophages.Elife. 2025 Jun 19;13:RP103714. doi: 10.7554/eLife.103714. Elife. 2025. PMID: 40536105 Free PMC article.
-
How lived experiences of illness trajectories, burdens of treatment, and social inequalities shape service user and caregiver participation in health and social care: a theory-informed qualitative evidence synthesis.Health Soc Care Deliv Res. 2025 Jun;13(24):1-120. doi: 10.3310/HGTQ8159. Health Soc Care Deliv Res. 2025. PMID: 40548558
-
Home treatment for mental health problems: a systematic review.Health Technol Assess. 2001;5(15):1-139. doi: 10.3310/hta5150. Health Technol Assess. 2001. PMID: 11532236
Cited by
-
Navigating contradictions in enteric chemotactic stimuli.Elife. 2025 Aug 11;14:RP106261. doi: 10.7554/eLife.106261. Elife. 2025. PMID: 40788306 Free PMC article.
-
Bacterial amino acid chemotaxis: a widespread strategy with multiple physiological and ecological roles.J Bacteriol. 2024 Oct 24;206(10):e0030024. doi: 10.1128/jb.00300-24. Epub 2024 Sep 27. J Bacteriol. 2024. PMID: 39330213 Free PMC article. Review.
-
Navigating contradictions: Salmonella Typhimurium chemotaxis amidst conflicting stimuli of the intestinal environment.bioRxiv [Preprint]. 2025 May 22:2024.01.18.576330. doi: 10.1101/2024.01.18.576330. bioRxiv. 2025. Update in: Elife. 2025 Aug 11;14:RP106261. doi: 10.7554/eLife.106261. PMID: 38293242 Free PMC article. Updated. Preprint.
-
Effects of Trichomonas gallinae infection and diet on blood microbiome composition in european greenfinches (Chloris chloris).Front Physiol. 2025 Jun 5;16:1576833. doi: 10.3389/fphys.2025.1576833. eCollection 2025. Front Physiol. 2025. PMID: 40538755 Free PMC article.
-
Identification of clinically relevant profiles in colorectal cancer through integrated analysis of bacterial DNA and metabolome in serum.Front Immunol. 2025 Jul 18;16:1562416. doi: 10.3389/fimmu.2025.1562416. eCollection 2025. Front Immunol. 2025. PMID: 40755755 Free PMC article.
References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX : a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

