A genetic screen of transcription factors in the Drosophila melanogaster abdomen identifies novel pigmentation genes
- PMID: 38820091
- PMCID: PMC11373662
- DOI: 10.1093/g3journal/jkae097
A genetic screen of transcription factors in the Drosophila melanogaster abdomen identifies novel pigmentation genes
Abstract
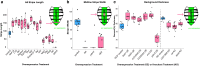
Gene regulatory networks specify the gene expression patterns needed for traits to develop. Differences in these networks can result in phenotypic differences between organisms. Although loss-of-function genetic screens can identify genes necessary for trait formation, gain-of-function screens can overcome genetic redundancy and identify loci whose expression is sufficient to alter trait formation. Here, we leveraged transgenic lines from the Transgenic RNAi Project at Harvard Medical School to perform both gain- and loss-of-function CRISPR/Cas9 screens for abdominal pigmentation phenotypes. We identified measurable effects on pigmentation patterns in the Drosophila melanogaster abdomen for 21 of 55 transcription factors in gain-of-function experiments and 7 of 16 tested by loss-of-function experiments. These included well-characterized pigmentation genes, such as bab1 and dsx, and transcription factors that had no known role in pigmentation, such as slp2. Finally, this screen was partially conducted by undergraduate students in a Genetics Laboratory course during the spring semesters of 2021 and 2022. We found this screen to be a successful model for student engagement in research in an undergraduate laboratory course that can be readily adapted to evaluate the effect of hundreds of genes on many different Drosophila traits, with minimal resources.
Keywords: Drosophila; CRISPR/Cas9; abdomen; development; gene regulation; pigmentation.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflict of interest The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
