Discovery and Validation of a Volatile Signature of Eosinophilic Airway Inflammation in Asthma
- PMID: 38820123
- PMCID: PMC11544360
- DOI: 10.1164/rccm.202310-1759OC
Discovery and Validation of a Volatile Signature of Eosinophilic Airway Inflammation in Asthma
Erratum in
-
Erratum: Discovery and Validation of a Volatile Signature of Eosinophilic Airway Inflammation in Asthma.Am J Respir Crit Care Med. 2025 Feb;211(2):296. doi: 10.1164/rccm.v211i2erratum1. Am J Respir Crit Care Med. 2025. PMID: 39887298 No abstract available.
Abstract
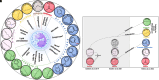
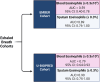
Rationale: Volatile organic compounds (VOCs) in asthmatic breath may be associated with sputum eosinophilia. We developed a volatile biomarker signature to predict sputum eosinophilia in asthma. Methods: VOCs emitted into the space above sputum samples (headspace) from patients with severe asthma (n = 36) were collected onto sorbent tubes and analyzed using thermal desorption gas chromatography-mass spectrometry (GC-MS). Elastic net regression identified stable VOCs associated with sputum eosinophilia ⩾ 3% and generated a volatile biomarker signature. This VOC signature was validated in breath samples from: 1) patients with acute asthma according to blood eosinophilia ⩾0.3 × 109cells/L or sputum eosinophilia of ⩾3% in the UK EMBER (East Midlands Breathomics Pathology Node) consortium (n = 65) and 2) U-BIOPRED-IMI (Unbiased Biomarkers in Prediction of Respiratory Disease Outcomes Innovative Medicines Initiative) consortium (n = 42). Breath samples were collected onto sorbent tubes (EMBER) or Tedlar bags (U-BIOPRED) and analyzed by GC-MS (GC × GC-MS for EMBER or GC-MS for U-BIOPRED). Measurements and Main Results: The in vitro headspace identified 19 VOCs associated with sputum eosinophilia, and the derived VOC signature yielded good diagnostic accuracy for sputum eosinophilia ⩾3% in headspace (area under the receiver operating characteristic curve [AUROC] 0.90; 95% confidence interval [CI], 0.80-0.99; P < 0.0001), correlated inversely with sputum eosinophil percentage (rs = -0.71; P < 0.0001), and outperformed fractional exhaled nitric oxide (AUROC 0.61; 95% CI, 0.35-0.86). Analysis of exhaled breath in replication cohorts yielded a VOC signature AUROC (95% CI) for acute asthma exacerbations of 0.89 (0.76-1.0) (EMBER cohort) with sputum eosinophilia and 0.90 (0.75-1.0) in U-BIOPRED, again outperforming fractional exhaled nitric oxide in U-BIOPRED (0.62 [0.33-0.90]). Conclusions: We have discovered and provided early-stage clinical validation of a volatile biomarker signature associated with eosinophilic airway inflammation. Further work is needed to translate our discovery using point-of-care clinical sensors.
Keywords: eosinophilic airway inflammation; severe asthma; volatile organic compound biomarkers.
Figures



Comment in
-
External Validation of Potential Breath Biomarkers for Asthma: A Step Forward Toward the Clinical Implementation of Breath Analysis.Am J Respir Crit Care Med. 2024 Nov 1;210(9):1069-1071. doi: 10.1164/rccm.202405-1033ED. Am J Respir Crit Care Med. 2024. PMID: 38924503 Free PMC article. No abstract available.
References
-
- Global Initiative for Asthma. 2022. www.ginasthma.org
-
- Castro M, Corren J, Pavord ID, Maspero J, Wenzel S, Rabe KF, et al. Dupilumab efficacy and safety in moderate-to-severe uncontrolled asthma. N Engl J Med . 2018;378:2486–2496. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

