Spinal inhibitory neurons degenerate before motor neurons and excitatory neurons in a mouse model of ALS
- PMID: 38820149
- PMCID: PMC11141618
- DOI: 10.1126/sciadv.adk3229
Spinal inhibitory neurons degenerate before motor neurons and excitatory neurons in a mouse model of ALS
Abstract
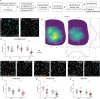
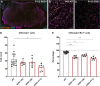
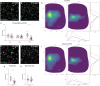
Amyotrophic lateral sclerosis (ALS) is characterized by the progressive loss of somatic motor neurons. A major focus has been directed to motor neuron intrinsic properties as a cause for degeneration, while less attention has been given to the contribution of spinal interneurons. In the present work, we applied multiplexing detection of transcripts and machine learning-based image analysis to investigate the fate of multiple spinal interneuron populations during ALS progression in the SOD1G93A mouse model. The analysis showed that spinal inhibitory interneurons are affected early in the disease, before motor neuron death, and are characterized by a slow progressive degeneration, while excitatory interneurons are affected later with a steep progression. Moreover, we report differential vulnerability within inhibitory and excitatory subpopulations. Our study reveals a strong interneuron involvement in ALS development with interneuron specific degeneration. These observations point to differential involvement of diverse spinal neuronal circuits that eventually may be determining motor neuron degeneration.
Figures






Similar articles
-
Nuclear Localization of Human SOD1 in Motor Neurons in Mouse Model and Patient Amyotrophic Lateral Sclerosis: Possible Links to Cholinergic Phenotype, NADPH Oxidase, Oxidative Stress, and DNA Damage.Int J Mol Sci. 2024 Aug 22;25(16):9106. doi: 10.3390/ijms25169106. Int J Mol Sci. 2024. PMID: 39201793 Free PMC article.
-
Gasdermin D is activated but does not drive neurodegeneration in SOD1G93A model of ALS: Implications for targeting pyroptosis.Neurobiol Dis. 2025 Oct 1;214:107048. doi: 10.1016/j.nbd.2025.107048. Epub 2025 Aug 6. Neurobiol Dis. 2025. PMID: 40759286 Free PMC article.
-
Desloratadine alleviates ALS-like pathology in hSOD1G93A mice via targeting 5HTR2A on activated spinal astrocytes.Acta Pharmacol Sin. 2024 May;45(5):926-944. doi: 10.1038/s41401-023-01223-2. Epub 2024 Jan 29. Acta Pharmacol Sin. 2024. PMID: 38286832 Free PMC article.
-
Proteostatic imbalance and protein spreading in amyotrophic lateral sclerosis.EMBO J. 2021 May 17;40(10):e106389. doi: 10.15252/embj.2020106389. Epub 2021 Mar 31. EMBO J. 2021. PMID: 33792056 Free PMC article. Review.
-
Treatment for sialorrhea (excessive saliva) in people with motor neuron disease/amyotrophic lateral sclerosis.Cochrane Database Syst Rev. 2022 May 20;5(5):CD006981. doi: 10.1002/14651858.CD006981.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593746 Free PMC article.
Cited by
-
Stabilization of V1 interneuron-motor neuron connectivity ameliorates motor phenotype in a mouse model of ALS.Nat Commun. 2024 Jun 7;15(1):4867. doi: 10.1038/s41467-024-48925-7. Nat Commun. 2024. PMID: 38849367 Free PMC article.
-
Potential contribution of spinal interneurons to the etiopathogenesis of amyotrophic lateral sclerosis.Front Neurosci. 2024 Jul 18;18:1434404. doi: 10.3389/fnins.2024.1434404. eCollection 2024. Front Neurosci. 2024. PMID: 39091344 Free PMC article. Review.
-
Spinal V1 inhibitory interneuron clades differ in birthdate, projections to motoneurons, and heterogeneity.Elife. 2024 Nov 28;13:RP95172. doi: 10.7554/eLife.95172. Elife. 2024. PMID: 39607843 Free PMC article.
-
The motor neuron m6A repertoire governs neuronal homeostasis and FTO inhibition mitigates ALS symptom manifestation.Nat Commun. 2025 Apr 30;16(1):4063. doi: 10.1038/s41467-025-59117-2. Nat Commun. 2025. PMID: 40307231 Free PMC article.
-
Synaptic imbalance and increased inhibition impair motor function in SMA.bioRxiv [Preprint]. 2024 Sep 1:2024.08.30.610545. doi: 10.1101/2024.08.30.610545. bioRxiv. 2024. PMID: 39257773 Free PMC article. Preprint.
References
-
- Kiehn O., Locomotor circuits in the mammalian spinal cord. Annu. Rev. Neurosci. 29, 279–306 (2006). - PubMed
-
- Briscoe J., Pierani A., Jessell T. M., Ericson J., A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101, 435–445 (2000). - PubMed
-
- Grillner S., El Manira A., The intrinsic operation of the networks that make us locomote. Curr. Opin. Neurobiol. 31, 244–249 (2015). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

