The KAT module of the SAGA complex maintains the oncogenic gene expression program in MYCN- amplified neuroblastoma
- PMID: 38820154
- PMCID: PMC11141635
- DOI: 10.1126/sciadv.adm9449
The KAT module of the SAGA complex maintains the oncogenic gene expression program in MYCN- amplified neuroblastoma
Abstract
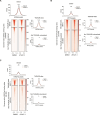
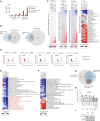
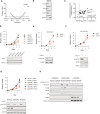
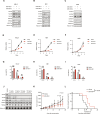
Pediatric cancers are frequently driven by genomic alterations that result in aberrant transcription factor activity. Here, we used functional genomic screens to identify multiple genes within the transcriptional coactivator Spt-Ada-Gcn5-acetyltransferase (SAGA) complex as selective dependencies for MYCN-amplified neuroblastoma, a disease of dysregulated development driven by an aberrant oncogenic transcriptional program. We characterized the DNA recruitment sites of the SAGA complex in neuroblastoma and the consequences of loss of SAGA complex lysine acetyltransferase (KAT) activity on histone acetylation and gene expression. We demonstrate that loss of SAGA complex KAT activity is associated with reduced MYCN binding on chromatin, suppression of MYC/MYCN gene expression programs, and impaired cell cycle progression. Further, we showed that the SAGA complex is pharmacologically targetable in vitro and in vivo with a KAT2A/KAT2B proteolysis targeting chimeric. Our findings expand our understanding of the histone-modifying complexes that maintain the oncogenic transcriptional state in this disease and suggest therapeutic potential for inhibitors of SAGA KAT activity in MYCN-amplified neuroblastoma.
Figures




References
-
- Yokoyama A., Somervaille T. C. P., Smith K. S., Rozenblatt-Rosen O., Meyerson M., Cleary M. L., The Menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell 123, 207–218 (2005). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

