Multimodal MALDI imaging mass spectrometry for improved diagnosis of melanoma
- PMID: 38820337
- PMCID: PMC11142536
- DOI: 10.1371/journal.pone.0304709
Multimodal MALDI imaging mass spectrometry for improved diagnosis of melanoma
Abstract
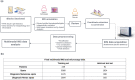
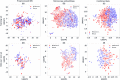
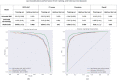
Imaging mass spectrometry (IMS) provides promising avenues to augment histopathological investigation with rich spatio-molecular information. We have previously developed a classification model to differentiate melanoma from nevi lesions based on IMS protein data, a task that is challenging solely by histopathologic evaluation. Most IMS-focused studies collect microscopy in tandem with IMS data, but this microscopy data is generally omitted in downstream data analysis. Microscopy, nevertheless, forms the basis for traditional histopathology and thus contains invaluable morphological information. In this work, we developed a multimodal classification pipeline that uses deep learning, in the form of a pre-trained artificial neural network, to extract the meaningful morphological features from histopathological images, and combine it with the IMS data. To test whether this deep learning-based classification strategy can improve on our previous results in classification of melanocytic neoplasia, we utilized MALDI IMS data with collected serial H&E stained sections for 331 patients, and compared this multimodal classification pipeline to classifiers using either exclusively microscopy or IMS data. The multimodal pipeline achieved the best performance, with ROC-AUCs of 0.968 vs. 0.938 vs. 0.931 for the multimodal, unimodal microscopy and unimodal IMS pipelines respectively. Due to the use of a pre-trained network to perform the morphological feature extraction, this pipeline does not require any training on large amounts of microscopy data. As such, this framework can be readily applied to improve classification performance in other experimental settings where microscopy data is acquired in tandem with IMS experiments.
Copyright: © 2024 Zhang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
JLM, NHP, SN, RMC, JLN, and JR disclose a financial interest in Fron- tier Diagnostics, LLC (FDx). FDx has issued and pending patent appli- cations in the US Patent Office that include part of the methods described in this paper. NV and MC, principals of Aspect Analytics NV, are paid consultants and provide services to FDx. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures


Similar articles
-
Diagnosis of melanoma by imaging mass spectrometry: Development and validation of a melanoma prediction model.J Cutan Pathol. 2021 Dec;48(12):1455-1462. doi: 10.1111/cup.14083. Epub 2021 Jul 2. J Cutan Pathol. 2021. PMID: 34151458 Free PMC article.
-
Exploring three-dimensional matrix-assisted laser desorption/ionization imaging mass spectrometry data: three-dimensional spatial segmentation of mouse kidney.Anal Chem. 2012 Jul 17;84(14):6079-87. doi: 10.1021/ac300673y. Epub 2012 Jul 5. Anal Chem. 2012. PMID: 22720760
-
Utility of imaging mass spectrometry (IMS) by matrix-assisted laser desorption ionization (MALDI) on an ion trap mass spectrometer in the analysis of drugs and metabolites in biological tissues.J Pharmacol Toxicol Methods. 2007 May-Jun;55(3):279-88. doi: 10.1016/j.vascn.2006.11.004. Epub 2006 Dec 5. J Pharmacol Toxicol Methods. 2007. PMID: 17222568
-
Skin cancer classification via convolutional neural networks: systematic review of studies involving human experts.Eur J Cancer. 2021 Oct;156:202-216. doi: 10.1016/j.ejca.2021.06.049. Epub 2021 Sep 8. Eur J Cancer. 2021. PMID: 34509059
-
Mass microscopy: high-resolution imaging mass spectrometry.J Electron Microsc (Tokyo). 2011;60(1):47-56. doi: 10.1093/jmicro/dfq079. Epub 2010 Nov 24. J Electron Microsc (Tokyo). 2011. PMID: 21109523 Review.
Cited by
-
Chemical imaging delineates Aβ plaque polymorphism across the Alzheimer's disease spectrum.Nat Commun. 2025 Apr 24;16(1):3889. doi: 10.1038/s41467-025-59085-7. Nat Commun. 2025. PMID: 40274785 Free PMC article.
-
MALDI imaging mass spectrometry differentiates basal cell carcinoma from trichoblastoma and trichoepithelioma: A proof of principle study.PLoS One. 2025 May 12;20(5):e0323475. doi: 10.1371/journal.pone.0323475. eCollection 2025. PLoS One. 2025. PMID: 40354485 Free PMC article.
References
-
- Available from: https://www.cancer.org/cancer/melanoma-skin-cancer/about/key-statistics..... Date of access: 2022-11-24.
-
- Veenhuizen KC, De Wit PE, Mooi WJ, Scheffer E, Verbeek AL, Ruiter DJ. Quality assessment by expert opinion in melanoma pathology: experience of the pathology panel of the Dutch Melanoma Working Party. J Pathol. 1997;182(3):266–272. doi: 10.1002/(SICI)1096-9896(199707)182:3<266::AID-PATH812>3.0.CO;2-# - DOI - PubMed
-
- Zembowicz A, Scolyer RA. Nevus/melanocytoma/melanoma: an emerging paradigm for classification of melanocytic neoplasms? Arch Pathol Lab Med. 2011;135(3):300–306. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

