Modeling spatial evolution of multi-drug resistance under drug environmental gradients
- PMID: 38820350
- PMCID: PMC11142541
- DOI: 10.1371/journal.pcbi.1012098
Modeling spatial evolution of multi-drug resistance under drug environmental gradients
Abstract
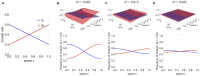
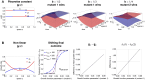
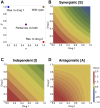
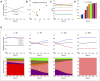
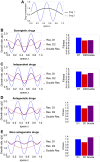
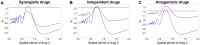
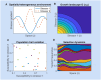
Multi-drug combinations to treat bacterial populations are at the forefront of approaches for infection control and prevention of antibiotic resistance. Although the evolution of antibiotic resistance has been theoretically studied with mathematical population dynamics models, extensions to spatial dynamics remain rare in the literature, including in particular spatial evolution of multi-drug resistance. In this study, we propose a reaction-diffusion system that describes the multi-drug evolution of bacteria based on a drug-concentration rescaling approach. We show how the resistance to drugs in space, and the consequent adaptation of growth rate, is governed by a Price equation with diffusion, integrating features of drug interactions and collateral resistances or sensitivities to the drugs. We study spatial versions of the model where the distribution of drugs is homogeneous across space, and where the drugs vary environmentally in a piecewise-constant, linear and nonlinear manner. Although in many evolution models, per capita growth rate is a natural surrogate for fitness, in spatially-extended, potentially heterogeneous habitats, fitness is an emergent property that potentially reflects additional complexities, from boundary conditions to the specific spatial variation of growth rates. Applying concepts from perturbation theory and reaction-diffusion equations, we propose an analytical metric for characterization of average mutant fitness in the spatial system based on the principal eigenvalue of our linear problem, λ1. This enables an accurate translation from drug spatial gradients and mutant antibiotic susceptibility traits to the relative advantage of each mutant across the environment. Our approach allows one to predict the precise outcomes of selection among mutants over space, ultimately from comparing their λ1 values, which encode a critical interplay between growth functions, movement traits, habitat size and boundary conditions. Such mathematical understanding opens new avenues for multi-drug therapeutic optimization.
Copyright: © 2024 Freire et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Update of
-
Modeling spatial evolution of multi-drug resistance under drug environmental gradients.bioRxiv [Preprint]. 2023 Nov 17:2023.11.16.567447. doi: 10.1101/2023.11.16.567447. bioRxiv. 2023. Update in: PLoS Comput Biol. 2024 May 31;20(5):e1012098. doi: 10.1371/journal.pcbi.1012098. PMID: 38014279 Free PMC article. Updated. Preprint.
Similar articles
-
Modeling spatial evolution of multi-drug resistance under drug environmental gradients.bioRxiv [Preprint]. 2023 Nov 17:2023.11.16.567447. doi: 10.1101/2023.11.16.567447. bioRxiv. 2023. Update in: PLoS Comput Biol. 2024 May 31;20(5):e1012098. doi: 10.1371/journal.pcbi.1012098. PMID: 38014279 Free PMC article. Updated. Preprint.
-
Price equation captures the role of drug interactions and collateral effects in the evolution of multidrug resistance.Elife. 2021 Jul 22;10:e64851. doi: 10.7554/eLife.64851. Elife. 2021. PMID: 34289932 Free PMC article.
-
Predictable properties of fitness landscapes induced by adaptational tradeoffs.Elife. 2020 May 19;9:e55155. doi: 10.7554/eLife.55155. Elife. 2020. PMID: 32423531 Free PMC article.
-
Evolutionary Trajectories to Antibiotic Resistance.Annu Rev Microbiol. 2017 Sep 8;71:579-596. doi: 10.1146/annurev-micro-090816-093813. Epub 2017 Jul 11. Annu Rev Microbiol. 2017. PMID: 28697667 Review.
-
Evolution of Drug Resistance in Bacteria.Adv Exp Med Biol. 2016;915:49-67. doi: 10.1007/978-3-319-32189-9_5. Adv Exp Med Biol. 2016. PMID: 27193537 Review.
Cited by
-
Deciphering and steering population-level response under spatial drug heterogeneity on microhabitat structures.bioRxiv [Preprint]. 2025 Jun 17:2025.02.13.638200. doi: 10.1101/2025.02.13.638200. bioRxiv. 2025. PMID: 40027692 Free PMC article. Preprint.
-
Phage-mediated peripheral kill-the-winner facilitates the maintenance of costly antibiotic resistance.Nat Commun. 2025 Jul 1;16(1):5839. doi: 10.1038/s41467-025-61055-y. Nat Commun. 2025. PMID: 40592899 Free PMC article.
-
Linking spatial drug heterogeneity to microbial growth dynamics in theory and experiment.bioRxiv [Preprint]. 2024 Nov 28:2024.11.21.624783. doi: 10.1101/2024.11.21.624783. bioRxiv. 2024. PMID: 39605592 Free PMC article. Preprint.
References
-
- Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, et al.. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. The Lancet infectious diseases. 2019;19(1):56–66. doi: 10.1016/S1473-3099(18)30605-4 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

