A pseudoautosomal glycosylation disorder prompts the revision of dolichol biosynthesis
- PMID: 38821050
- PMCID: PMC11250103
- DOI: 10.1016/j.cell.2024.04.041
A pseudoautosomal glycosylation disorder prompts the revision of dolichol biosynthesis
Erratum in
-
A pseudoautosomal glycosylation disorder prompts the revision of dolichol biosynthesis.Cell. 2024 Jul 11;187(14):3784. doi: 10.1016/j.cell.2024.06.004. Epub 2024 Jun 12. Cell. 2024. PMID: 38870945 Free PMC article. No abstract available.
Abstract
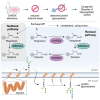
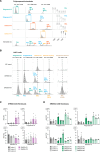
Dolichol is a lipid critical for N-glycosylation as a carrier for activated sugars and nascent oligosaccharides. It is commonly thought to be directly produced from polyprenol by the enzyme SRD5A3. Instead, we found that dolichol synthesis requires a three-step detour involving additional metabolites, where SRD5A3 catalyzes only the second reaction. The first and third steps are performed by DHRSX, whose gene resides on the pseudoautosomal regions of the X and Y chromosomes. Accordingly, we report a pseudoautosomal-recessive disease presenting as a congenital disorder of glycosylation in patients with missense variants in DHRSX (DHRSX-CDG). Of note, DHRSX has a unique dual substrate and cofactor specificity, allowing it to act as a NAD+-dependent dehydrogenase and as a NADPH-dependent reductase in two non-consecutive steps. Thus, our work reveals unexpected complexity in the terminal steps of dolichol biosynthesis. Furthermore, we provide insights into the mechanism by which dolichol metabolism defects contribute to disease.
Keywords: N-glycosylation; congenital disorders of glycosylation; dolichal; dolichol; lipid droplets; polyisoprenoids; polyprenal; polyprenol; pseudoautosomal region.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures











References
-
- Shridas P., Rush J.S., Waechter C.J. Identification and characterization of a cDNA encoding a long-chain cis-isoprenyltranferase involved in dolichyl monophosphate biosynthesis in the ER of brain cells. Biochem. Biophys. Res. Commun. 2003;312:1349–1356. doi: 10.1016/j.bbrc.2003.11.065. - DOI - PubMed
-
- Cantagrel V., Lefeber D.J., Ng B.G., Guan Z., Silhavy J.L., Bielas S.L., Lehle L., Hombauer H., Adamowicz M., Swiezewska E., et al. SRD5A3 Is Required for Converting Polyprenol to Dolichol and Is Mutated in a Congenital Glycosylation Disorder. Cell. 2010;142:203–217. doi: 10.1016/j.cell.2010.06.001. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

