Universal paramyxovirus vaccine design by stabilizing regions involved in structural transformation of the fusion protein
- PMID: 38821950
- PMCID: PMC11143371
- DOI: 10.1038/s41467-024-48059-w
Universal paramyxovirus vaccine design by stabilizing regions involved in structural transformation of the fusion protein
Abstract
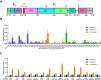
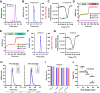
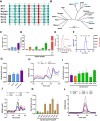
The Paramyxoviridae family encompasses medically significant RNA viruses, including human respiroviruses 1 and 3 (RV1, RV3), and zoonotic pathogens like Nipah virus (NiV). RV3, previously known as parainfluenza type 3, for which no vaccines or antivirals have been approved, causes respiratory tract infections in vulnerable populations. The RV3 fusion (F) protein is inherently metastable and will likely require prefusion (preF) stabilization for vaccine effectiveness. Here we used structure-based design to stabilize regions involved in structural transformation to generate a preF protein vaccine antigen with high expression and stability, and which, by stabilizing the coiled-coil stem region, does not require a heterologous trimerization domain. The preF candidate induces strong neutralizing antibody responses in both female naïve and pre-exposed mice and provides protection in a cotton rat challenge model (female). Despite the evolutionary distance of paramyxovirus F proteins, their structural transformation and local regions of instability are conserved, which allows successful transfer of stabilizing substitutions to the distant preF proteins of RV1 and NiV. This work presents a successful vaccine antigen design for RV3 and provides a toolbox for future paramyxovirus vaccine design and pandemic preparedness.
© 2024. The Author(s).
Conflict of interest statement
J.P.M.L., J.J., and M.J.G.B. are co-inventors on related vaccine patents. J.P.M.L., F.C., D.v.O., L.L., W.v.d.H., R.V., R.Z., L.v.d.F., J.J. and M.J.G.B. are employees of Janssen Vaccines & Prevention BV and may hold stock of Johnson and Johnson.
Figures





Similar articles
-
Efficacious human metapneumovirus vaccine based on AI-guided engineering of a closed prefusion trimer.Nat Commun. 2024 Jul 25;15(1):6270. doi: 10.1038/s41467-024-50659-5. Nat Commun. 2024. PMID: 39054318 Free PMC article.
-
Structure-based design of a quadrivalent fusion glycoprotein vaccine for human parainfluenza virus types 1-4.Proc Natl Acad Sci U S A. 2018 Nov 27;115(48):12265-12270. doi: 10.1073/pnas.1811980115. Epub 2018 Nov 12. Proc Natl Acad Sci U S A. 2018. PMID: 30420505 Free PMC article.
-
Enhanced Neutralizing Antibody Response Induced by Respiratory Syncytial Virus Prefusion F Protein Expressed by a Vaccine Candidate.J Virol. 2015 Sep;89(18):9499-510. doi: 10.1128/JVI.01373-15. Epub 2015 Jul 8. J Virol. 2015. PMID: 26157122 Free PMC article.
-
Packaging and Prefusion Stabilization Separately and Additively Increase the Quantity and Quality of Respiratory Syncytial Virus (RSV)-Neutralizing Antibodies Induced by an RSV Fusion Protein Expressed by a Parainfluenza Virus Vector.J Virol. 2016 Oct 14;90(21):10022-10038. doi: 10.1128/JVI.01196-16. Print 2016 Nov 1. J Virol. 2016. PMID: 27581977 Free PMC article.
-
Structure-Based Design of Nipah Virus Vaccines: A Generalizable Approach to Paramyxovirus Immunogen Development.Front Immunol. 2020 Jun 11;11:842. doi: 10.3389/fimmu.2020.00842. eCollection 2020. Front Immunol. 2020. PMID: 32595632 Free PMC article.
Cited by
-
A foldon-free prefusion F trimer vaccine for respiratory syncytial virus to reduce off-target immune responses.Nat Microbiol. 2024 Dec;9(12):3254-3267. doi: 10.1038/s41564-024-01860-1. Epub 2024 Nov 20. Nat Microbiol. 2024. PMID: 39567664 Free PMC article.
-
Zoonotic Paramyxoviruses: Evolution, Ecology, and Public Health Strategies in a Changing World.Viruses. 2024 Oct 29;16(11):1688. doi: 10.3390/v16111688. Viruses. 2024. Retraction in: Viruses. 2025 Jul 16;17(7):992. doi: 10.3390/v17070992. PMID: 39599803 Free PMC article. Retracted. Review.
-
Advancements in the genomic feature of Newcastle disease virus and the multifaceted roles of non-structural proteins V/W in viral replication and pathogenesis.Poult Sci. 2025 Jul 3;104(10):105527. doi: 10.1016/j.psj.2025.105527. Online ahead of print. Poult Sci. 2025. PMID: 40639003 Free PMC article. Review.
-
Design, Structure, and Immunogenicity of a Soluble Prefusion-stabilized EBV gB Antigen.bioRxiv [Preprint]. 2025 May 19:2025.05.19.654955. doi: 10.1101/2025.05.19.654955. bioRxiv. 2025. PMID: 40661529 Free PMC article. Preprint.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

