Targeting the autophagy-NAD axis protects against cell death in Niemann-Pick type C1 disease models
- PMID: 38821960
- PMCID: PMC11143325
- DOI: 10.1038/s41419-024-06770-y
Targeting the autophagy-NAD axis protects against cell death in Niemann-Pick type C1 disease models
Abstract
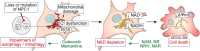
Impairment of autophagy leads to an accumulation of misfolded proteins and damaged organelles and has been implicated in plethora of human diseases. Loss of autophagy in actively respiring cells has also been shown to trigger metabolic collapse mediated by the depletion of nicotinamide adenine dinucleotide (NAD) pools, resulting in cell death. Here we found that the deficit in the autophagy-NAD axis underpins the loss of viability in cell models of a neurodegenerative lysosomal storage disorder, Niemann-Pick type C1 (NPC1) disease. Defective autophagic flux in NPC1 cells resulted in mitochondrial dysfunction due to impairment of mitophagy, leading to the depletion of both the reduced and oxidised forms of NAD as identified via metabolic profiling. Consequently, exhaustion of the NAD pools triggered mitochondrial depolarisation and apoptotic cell death. Our chemical screening identified two FDA-approved drugs, celecoxib and memantine, as autophagy activators which effectively restored autophagic flux, NAD levels, and cell viability of NPC1 cells. Of biomedical relevance, either pharmacological rescue of the autophagy deficiency or NAD precursor supplementation restored NAD levels and improved the viability of NPC1 patient fibroblasts and induced pluripotent stem cell (iPSC)-derived cortical neurons. Together, our findings identify the autophagy-NAD axis as a mechanism of cell death and a target for therapeutic interventions in NPC1 disease, with a potential relevance to other neurodegenerative disorders.
© 2024. The Author(s).
Conflict of interest statement
J.E.O. is an employee of The Procter & Gamble Company, USA. S. Sarkar is a scientific consultant for Ayin Tech LLC. V.I.K. is a Scientific Advisor for Longaevus Technologies.
Figures






Similar articles
-
Itraconazole and posaconazole, inhibitors of NPC1 sterol transport, act as pharmacological chaperones after washout.J Biol Chem. 2025 Jul;301(7):110370. doi: 10.1016/j.jbc.2025.110370. Epub 2025 Jun 16. J Biol Chem. 2025. PMID: 40516874 Free PMC article.
-
Myeloid cell-specific loss of NPC1 in mice recapitulates microgliosis and neurodegeneration in patients with Niemann-Pick type C disease.Sci Transl Med. 2024 Dec 4;16(776):eadl4616. doi: 10.1126/scitranslmed.adl4616. Epub 2024 Dec 4. Sci Transl Med. 2024. PMID: 39630885
-
LC3 Immunostaining in the Inferior Olivary Nuclei of Cats With Niemann-Pick Disease Type C1 Is Associated With Patterned Purkinje Cell Loss.J Neuropathol Exp Neurol. 2018 Mar 1;77(3):229-245. doi: 10.1093/jnen/nlx119. J Neuropathol Exp Neurol. 2018. PMID: 29346563 Free PMC article.
-
Biomarker Validation in NPC1: Foundations for Clinical Trials and Regulatory Alignment.J Inherit Metab Dis. 2025 Sep;48(5):e70075. doi: 10.1002/jimd.70075. J Inherit Metab Dis. 2025. PMID: 40814945 Free PMC article. Review.
-
Dysphagia as a risk factor for mortality in Niemann-Pick disease type C: systematic literature review and evidence from studies with miglustat.Orphanet J Rare Dis. 2012 Oct 6;7:76. doi: 10.1186/1750-1172-7-76. Orphanet J Rare Dis. 2012. PMID: 23039766 Free PMC article.
Cited by
-
Targeting endoplasmic reticulum stress-induced lymphatic dysfunction for mitigating bisphosphonate-related osteonecrosis.Clin Transl Med. 2024 Nov;14(11):e70082. doi: 10.1002/ctm2.70082. Clin Transl Med. 2024. PMID: 39521624 Free PMC article.
-
Metabolic checkpoints in immune cell reprogramming: rewiring immunometabolism for cancer therapy.Mol Cancer. 2025 Aug 2;24(1):210. doi: 10.1186/s12943-025-02407-6. Mol Cancer. 2025. PMID: 40753443 Free PMC article. Review.
-
Spatio-temporal processes in autophagosome-lysosome fusion.Med Rev (2021). 2025 Jun 19;5(4):297-317. doi: 10.1515/mr-2024-0095. eCollection 2025 Aug. Med Rev (2021). 2025. PMID: 40838105 Free PMC article. Review.
-
Current strategies in the management of dementia with lewy bodies and future directions based on disease pathophysiology.Mol Neurodegener. 2025 Jul 1;20(1):78. doi: 10.1186/s13024-025-00856-7. Mol Neurodegener. 2025. PMID: 40598239 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- ARUK-SRF2022A-006/Alzheimer's Research UK (ARUK)
- P2019-0004/LifeArc
- BB/R008167/2/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BH174490/RCUK | MRC | Medical Research Foundation
- 2016-17-0087/UK-India Education and Research Initiative (UKIERI)
- 109626/Z/15/Z/Wellcome Trust (Wellcome)
- BB/M023389/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- WT_/Wellcome Trust/United Kingdom
- 19J12969/MEXT | Japan Society for the Promotion of Science (JSPS)
- 1516ISSFFEL10/Wellcome Trust (Wellcome)
- Pathfinder 2324602/LifeArc
- RF1AG55549/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- C53309/A19702/Cancer Research UK (CRUK)
- 18KK0242/MEXT | Japan Society for the Promotion of Science (JSPS)
- RF1 AG055549/AG/NIA NIH HHS/United States
- BH174490/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Research Materials

