Cryo-EM structure of cadmium-bound human ABCB6
- PMID: 38822018
- PMCID: PMC11143254
- DOI: 10.1038/s42003-024-06377-1
Cryo-EM structure of cadmium-bound human ABCB6
Abstract
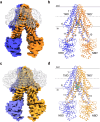
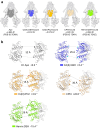
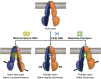
ATP-binding cassette transporter B6 (ABCB6), a protein essential for heme biosynthesis in mitochondria, also functions as a heavy metal efflux pump. Here, we present cryo-electron microscopy structures of human ABCB6 bound to a cadmium Cd(II) ion in the presence of antioxidant thiol peptides glutathione (GSH) and phytochelatin 2 (PC2) at resolutions of 3.2 and 3.1 Å, respectively. The overall folding of the two structures resembles the inward-facing apo state but with less separation between the two halves of the transporter. Two GSH molecules are symmetrically bound to the Cd(II) ion in a bent conformation, with the central cysteine protruding towards the metal. The N-terminal glutamate and C-terminal glycine of GSH do not directly interact with Cd(II) but contribute to neutralizing positive charges of the binding cavity by forming hydrogen bonds and van der Waals interactions with nearby residues. In the presence of PC2, Cd(II) binding to ABCB6 is similar to that observed with GSH, except that two cysteine residues of each PC2 molecule participate in Cd(II) coordination to form a tetrathiolate. Structural comparison of human ABCB6 and its homologous Atm-type transporters indicate that their distinct substrate specificity might be attributed to variations in the capping residues situated at the top of the substrate-binding cavity.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Cryo-electron microscopy structure of human ABCB6 transporter.Protein Sci. 2020 Dec;29(12):2363-2374. doi: 10.1002/pro.3960. Epub 2020 Oct 15. Protein Sci. 2020. PMID: 33007128 Free PMC article.
-
Structural Insights into Porphyrin Recognition by the Human ATP-Binding Cassette Transporter ABCB6.Mol Cells. 2022 Aug 31;45(8):575-587. doi: 10.14348/molcells.2022.0040. Epub 2022 Jul 28. Mol Cells. 2022. PMID: 35950458 Free PMC article.
-
W546 stacking disruption traps the human porphyrin transporter ABCB6 in an outward-facing transient state.Commun Biol. 2023 Sep 21;6(1):960. doi: 10.1038/s42003-023-05339-3. Commun Biol. 2023. PMID: 37735522 Free PMC article.
-
ABCB6, an ABC Transporter Impacting Drug Response and Disease.AAPS J. 2017 Nov 30;20(1):8. doi: 10.1208/s12248-017-0165-6. AAPS J. 2017. PMID: 29192381 Free PMC article. Review.
-
Cryo-EM of ABC transporters: an ice-cold solution to everything?FEBS Lett. 2020 Dec;594(23):3776-3789. doi: 10.1002/1873-3468.13989. FEBS Lett. 2020. PMID: 33156959 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

